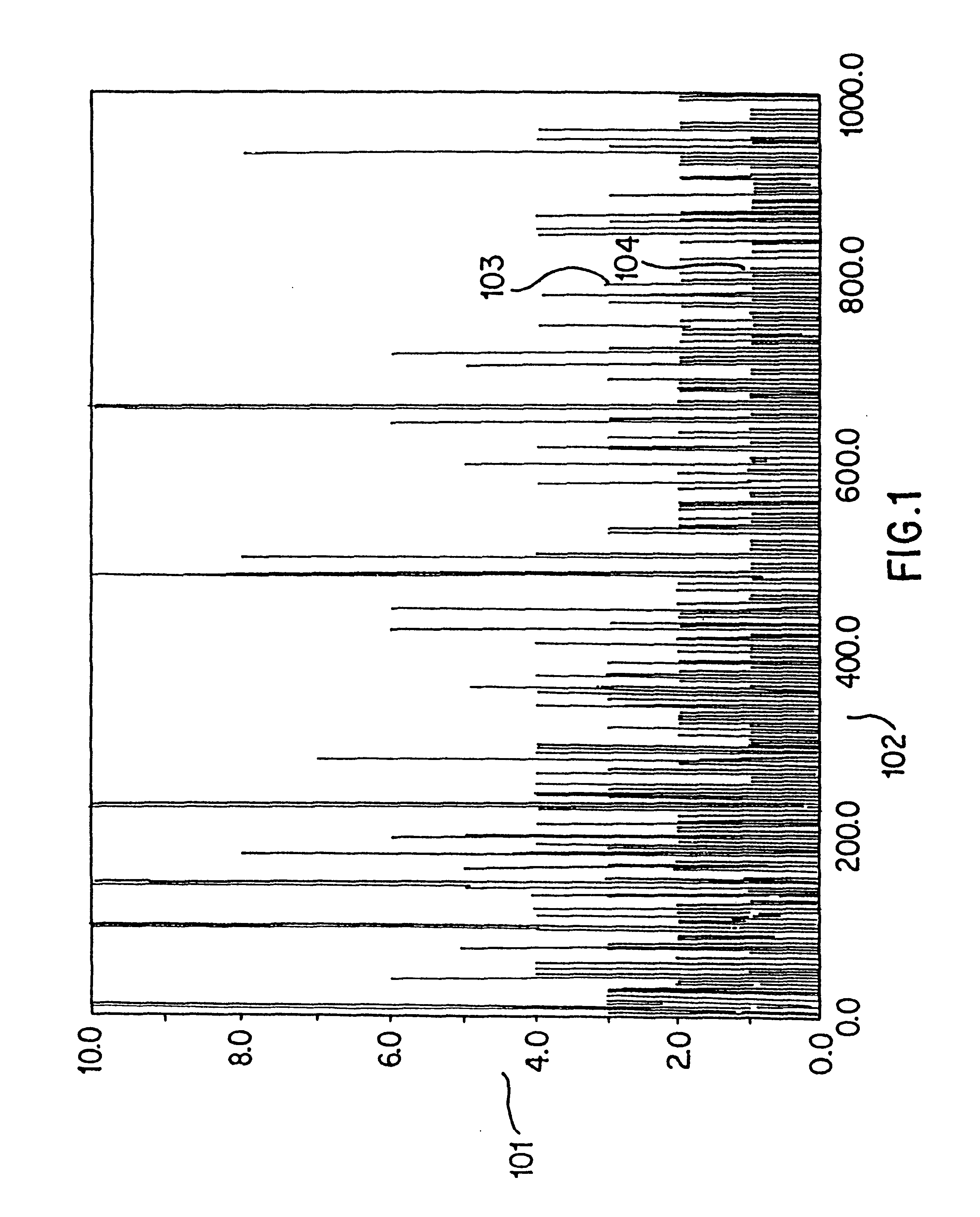
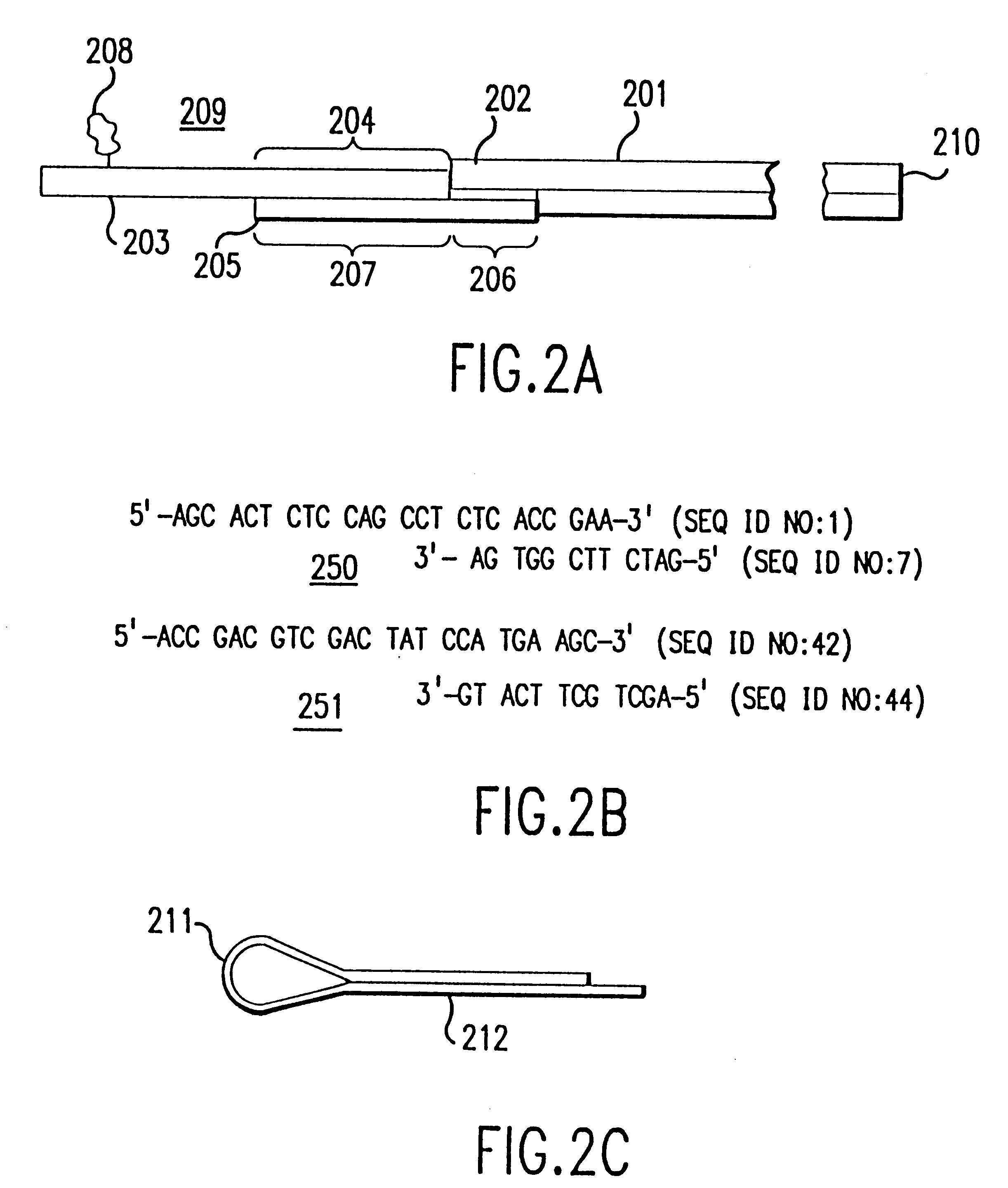
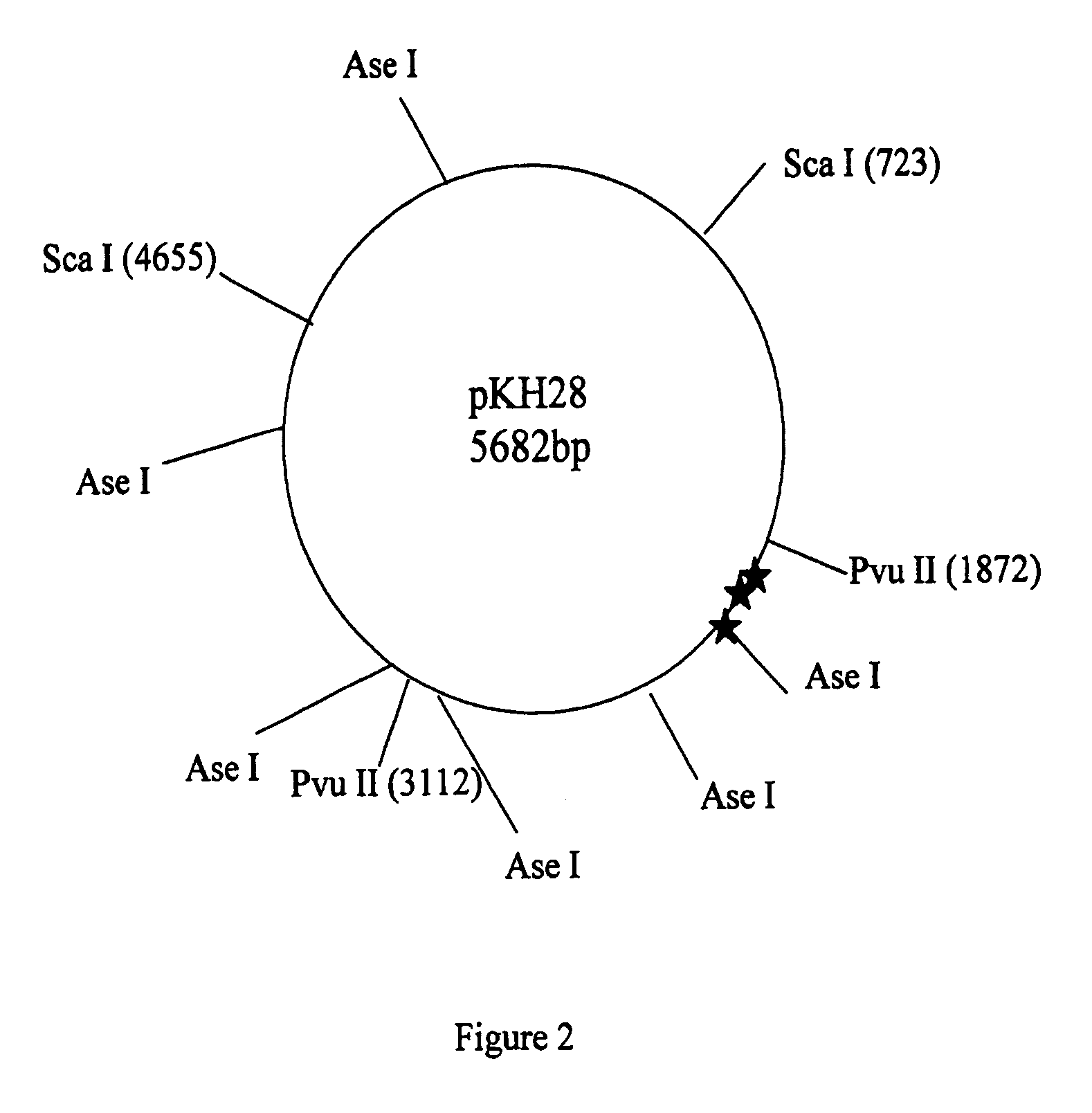
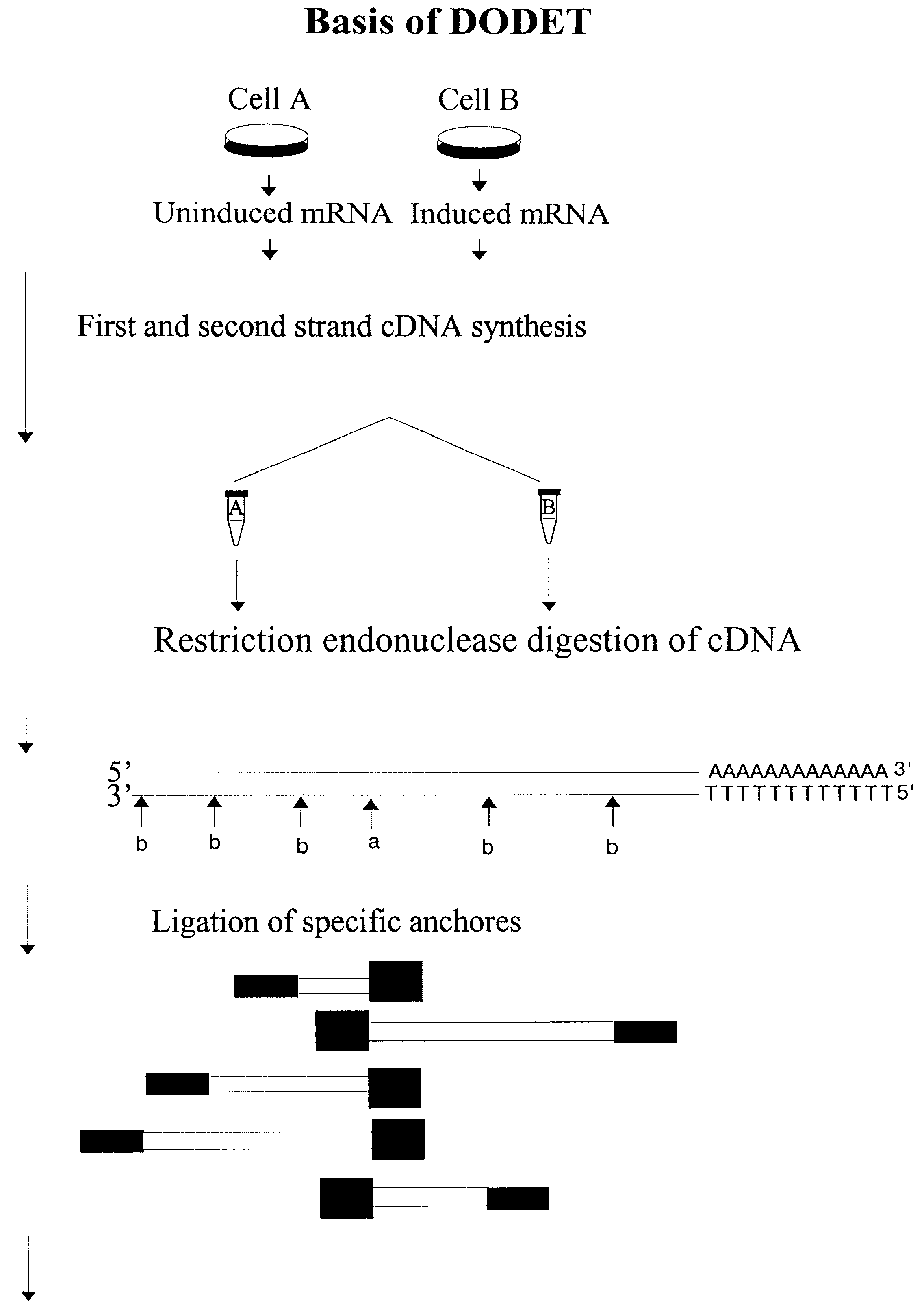
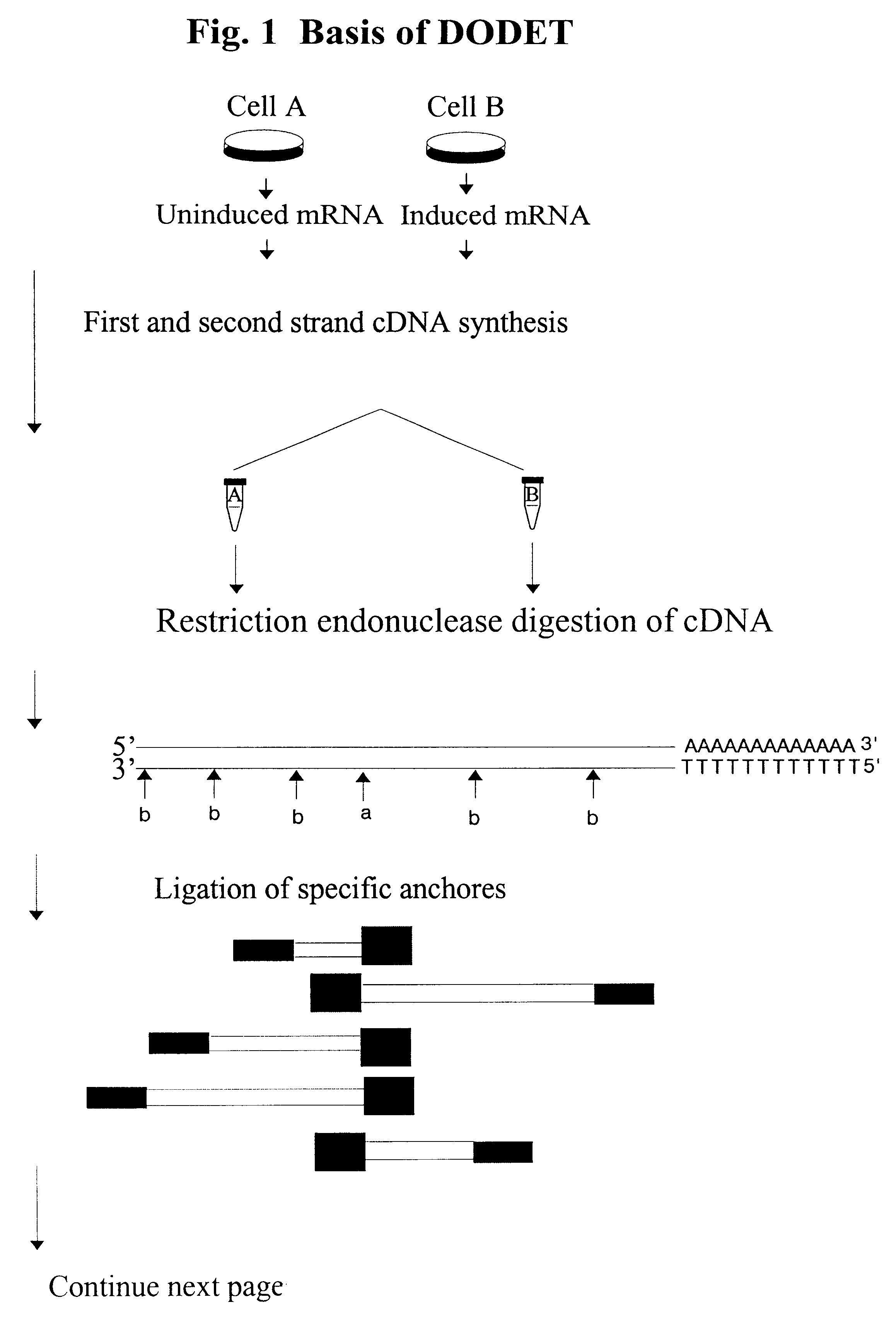
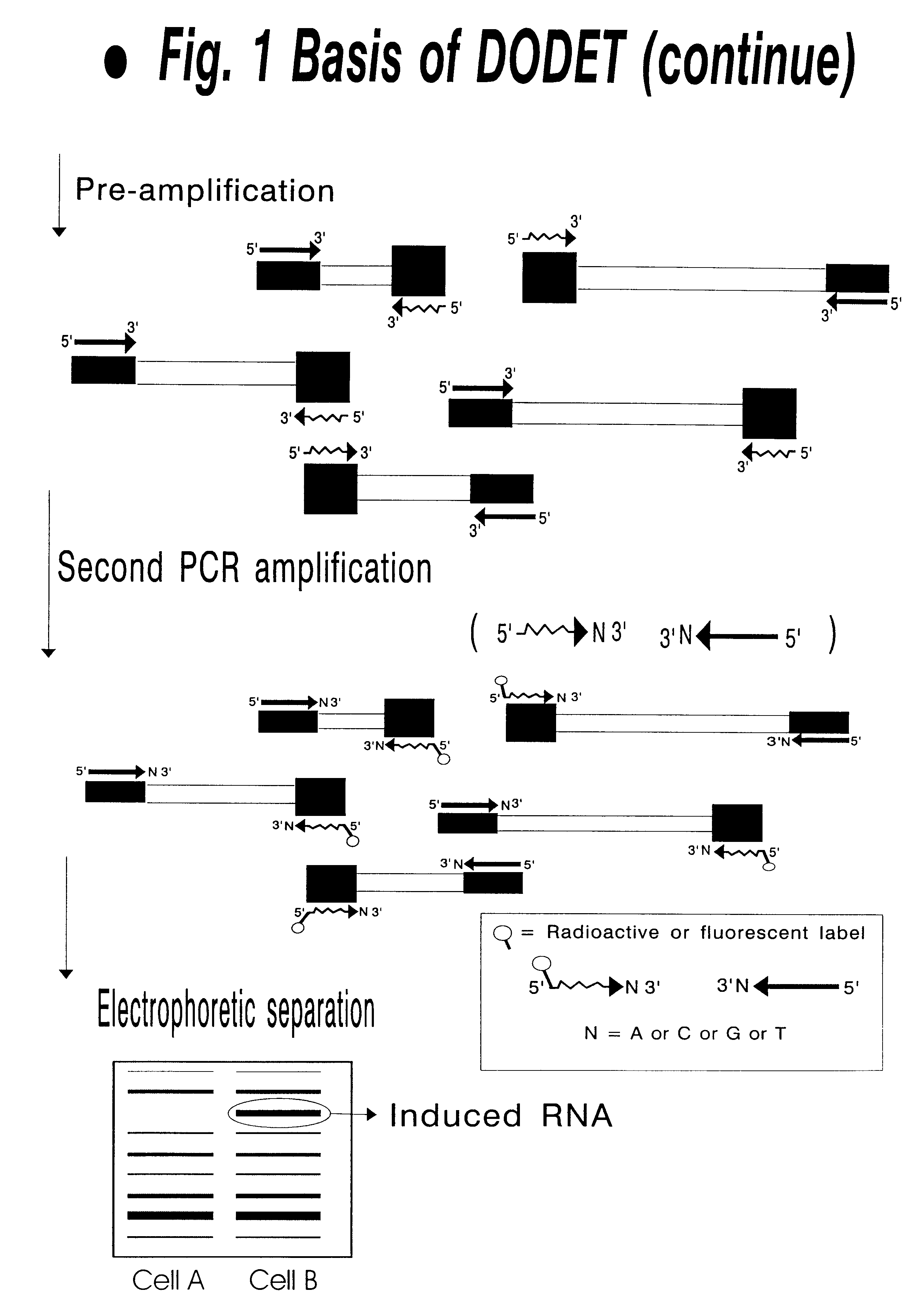
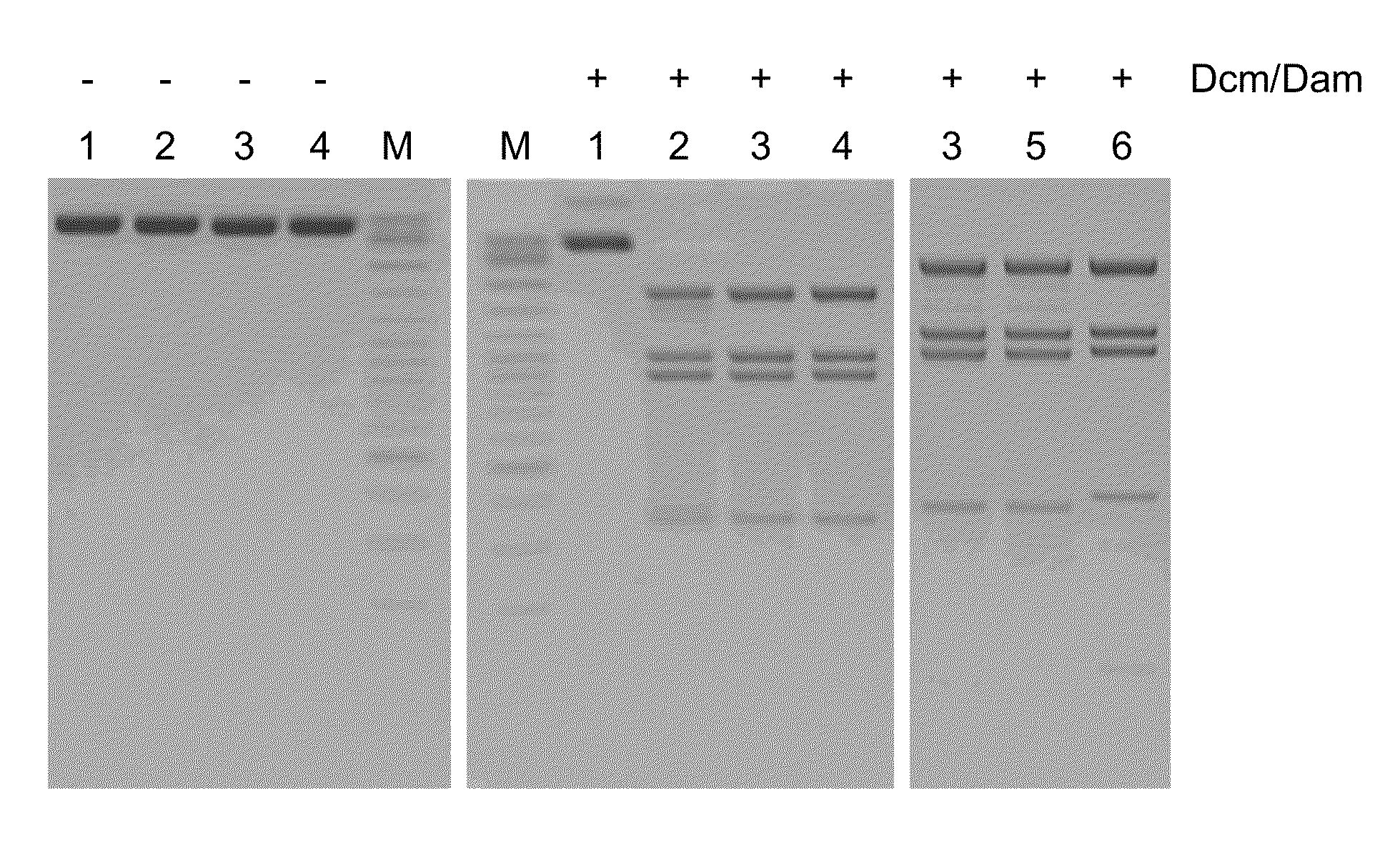
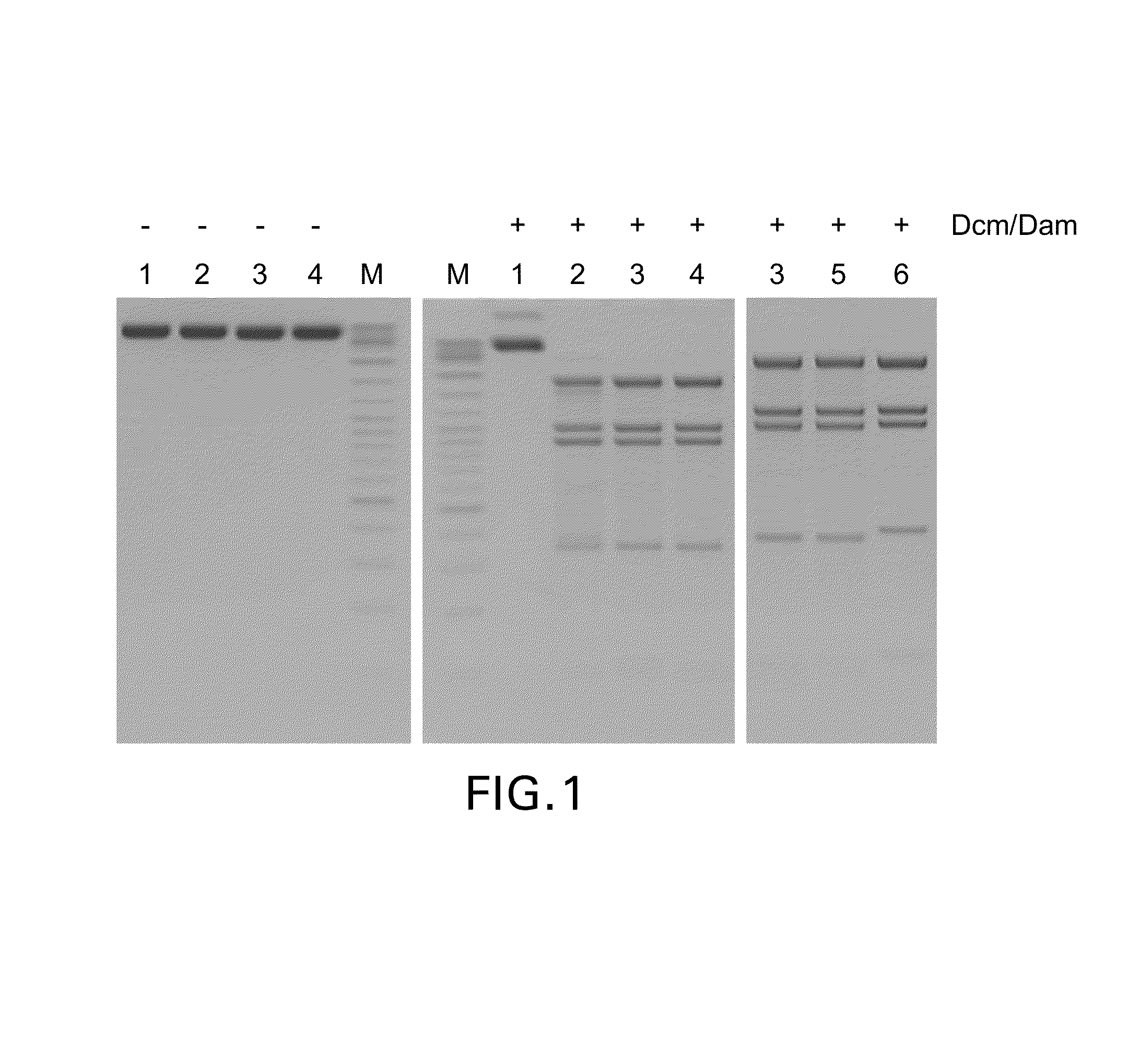
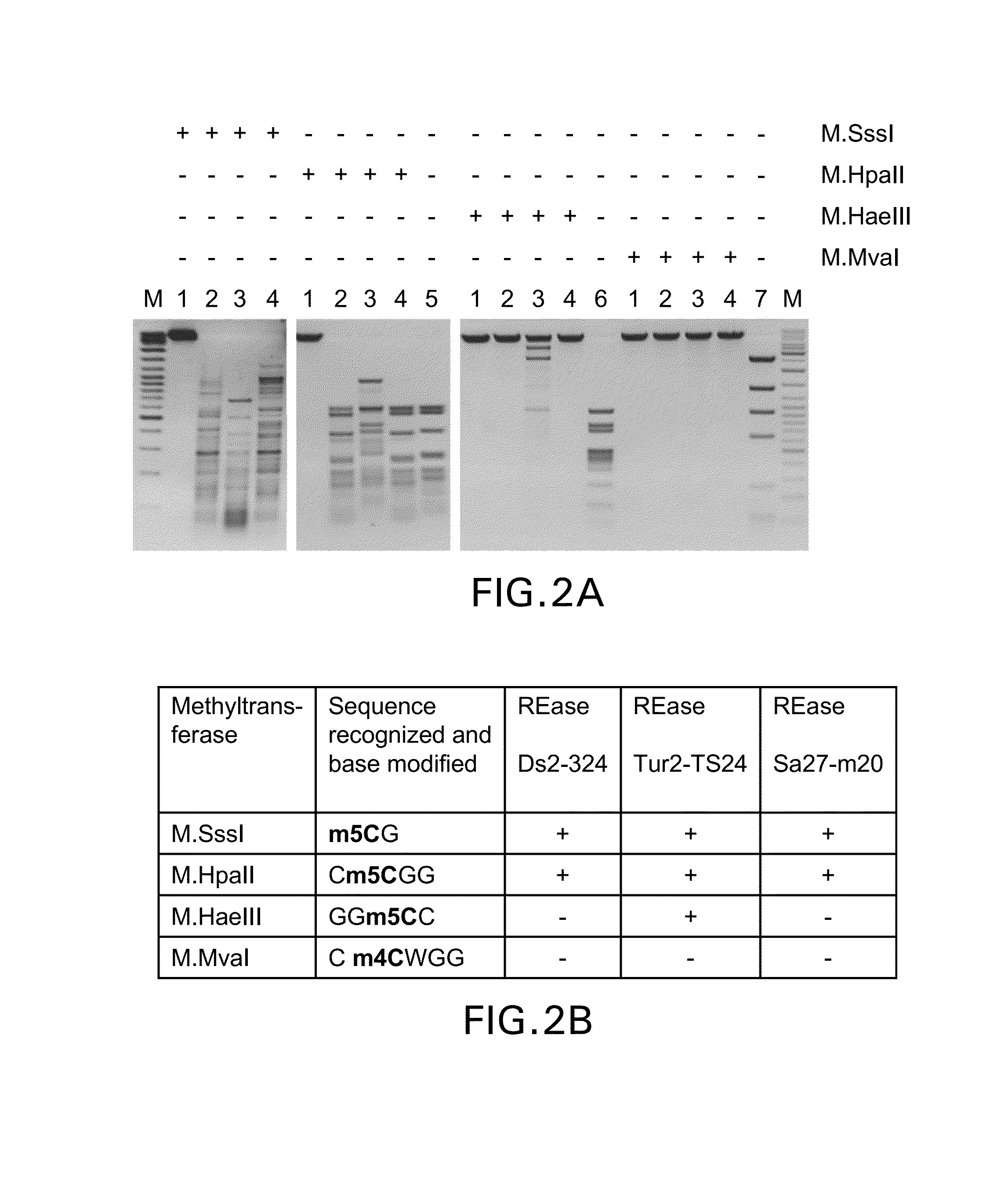
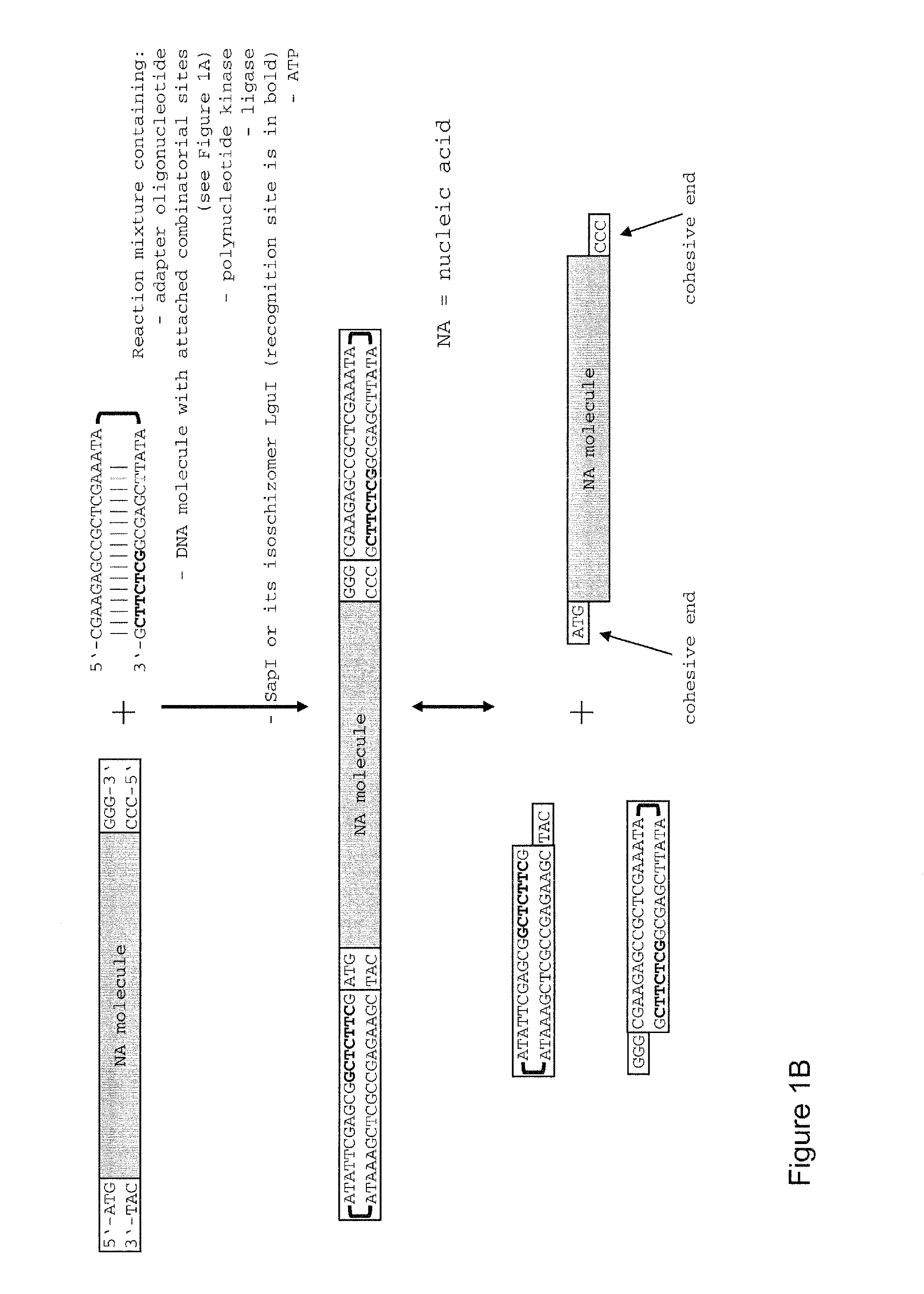
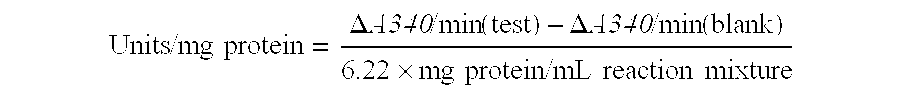Patents
Literature
101 results about "SmaI restriction endonuclease" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
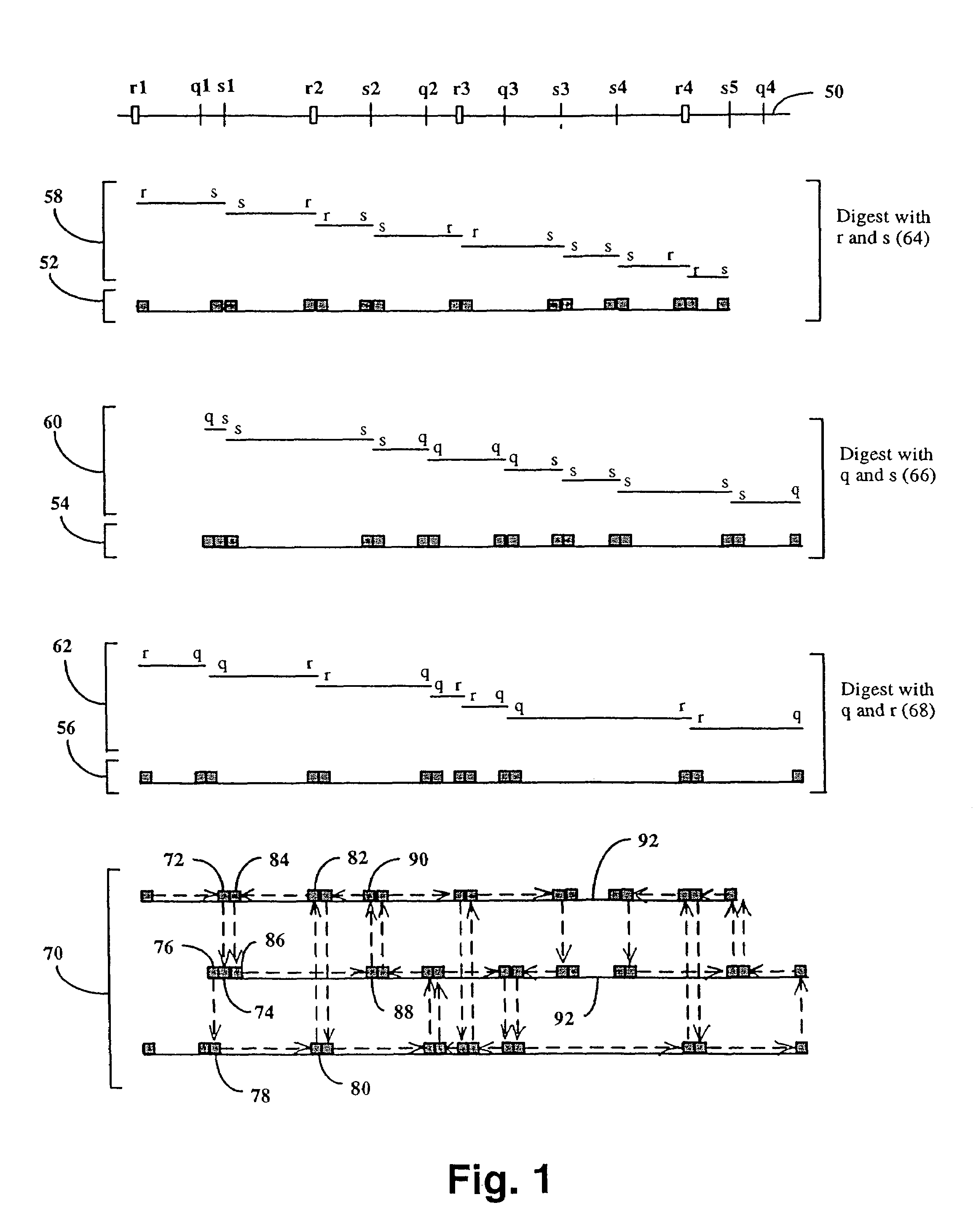
Method and compositions for ordering restriction fragments
InactiveUS7598035B2High density physicalPrecise positioningSugar derivativesMicrobiological testing/measurementPhysical MapsNucleotide sequencing
The invention provides a method for constructing a high resolution physical map of a polynucleotide. In accordance with the invention, nucleotide sequences are determined at the ends of restriction fragments produced by a plurality of digestions with a plurality of combinations of restriction endonucleases so that a pair of nucleotide sequences is obtained for each restriction fragment. A physical map of the polynucleotide is constructed by ordering the pairs of sequences by matching the identical sequences among the pairs.
Owner:ILLUMINA INC
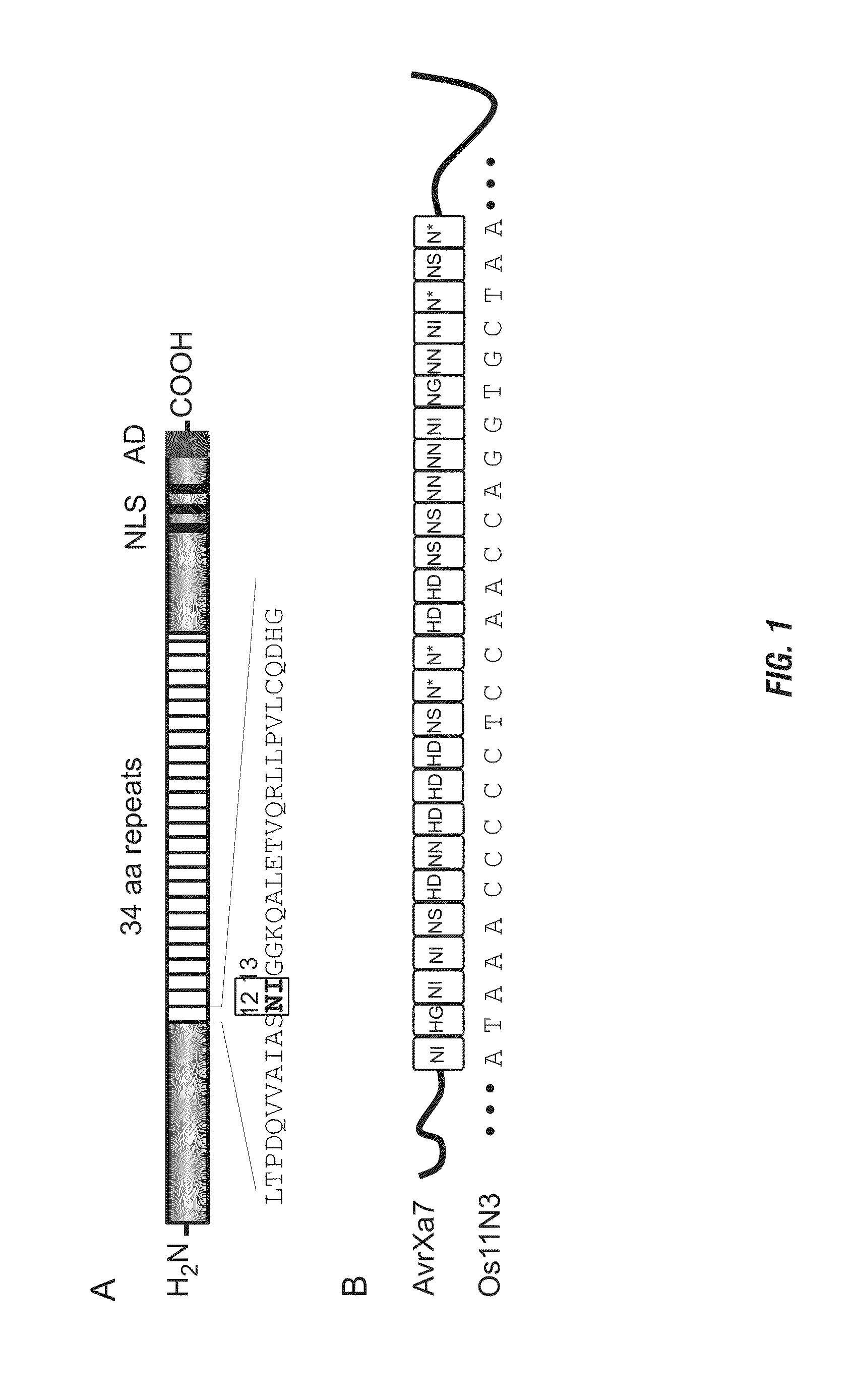
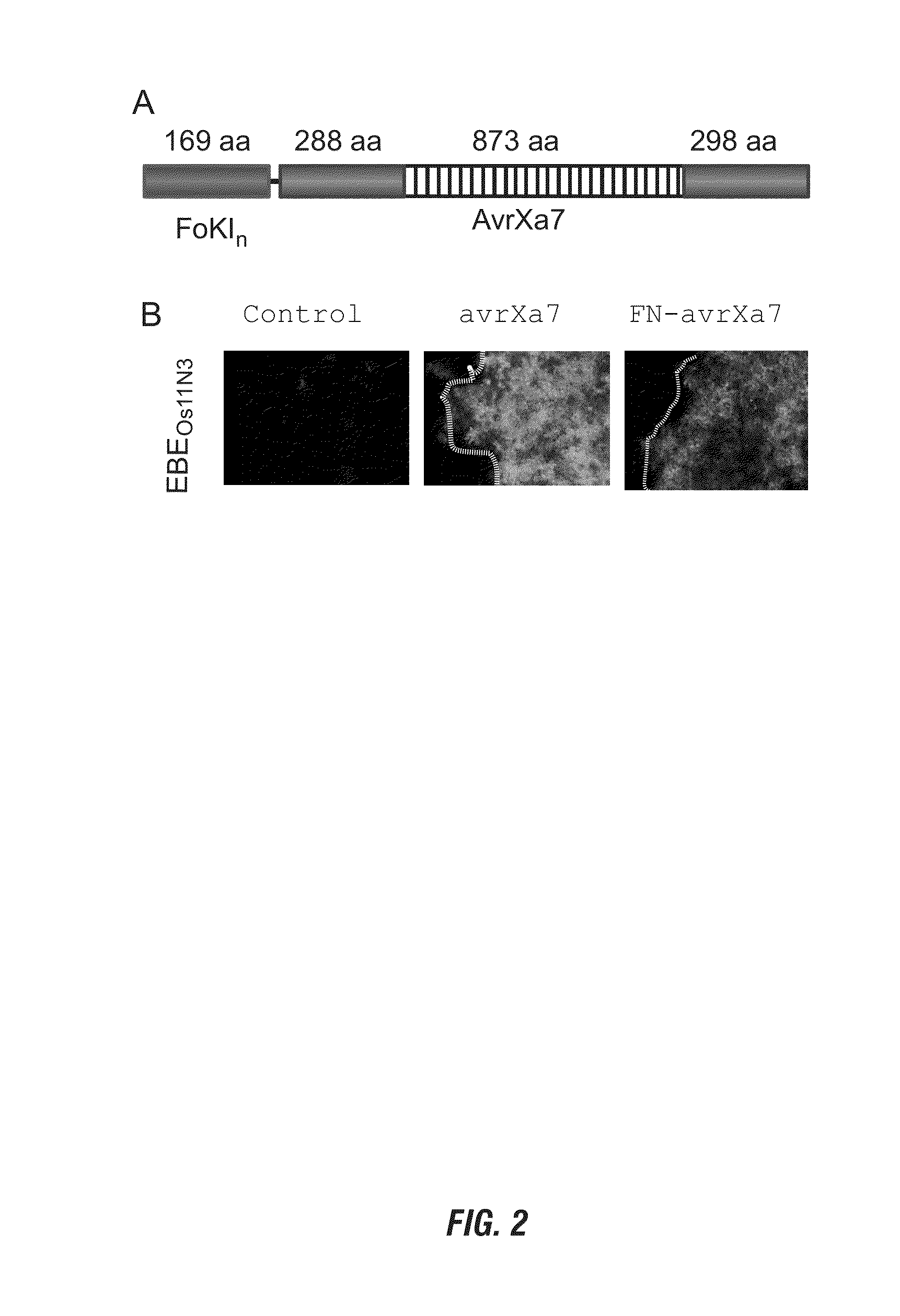
Nuclease activity of tal effector and foki fusion protein
The present invention provides compositions and methods for targeted cleavage of cellular chromatin in a region of interest and / or homologous recombination at a predetermined site in cells. Compositions include fusion polypeptides comprising a TAL effector binding domain and a cleavage domain. The cleavage domain can be from any endonuclease. In certain embodiments, the endonuclease is a Type IIS restriction endonuclease. In further embodiments, the Type IIS restriction endonuclease is FokI.
Owner:IOWA STATE UNIV RES FOUND
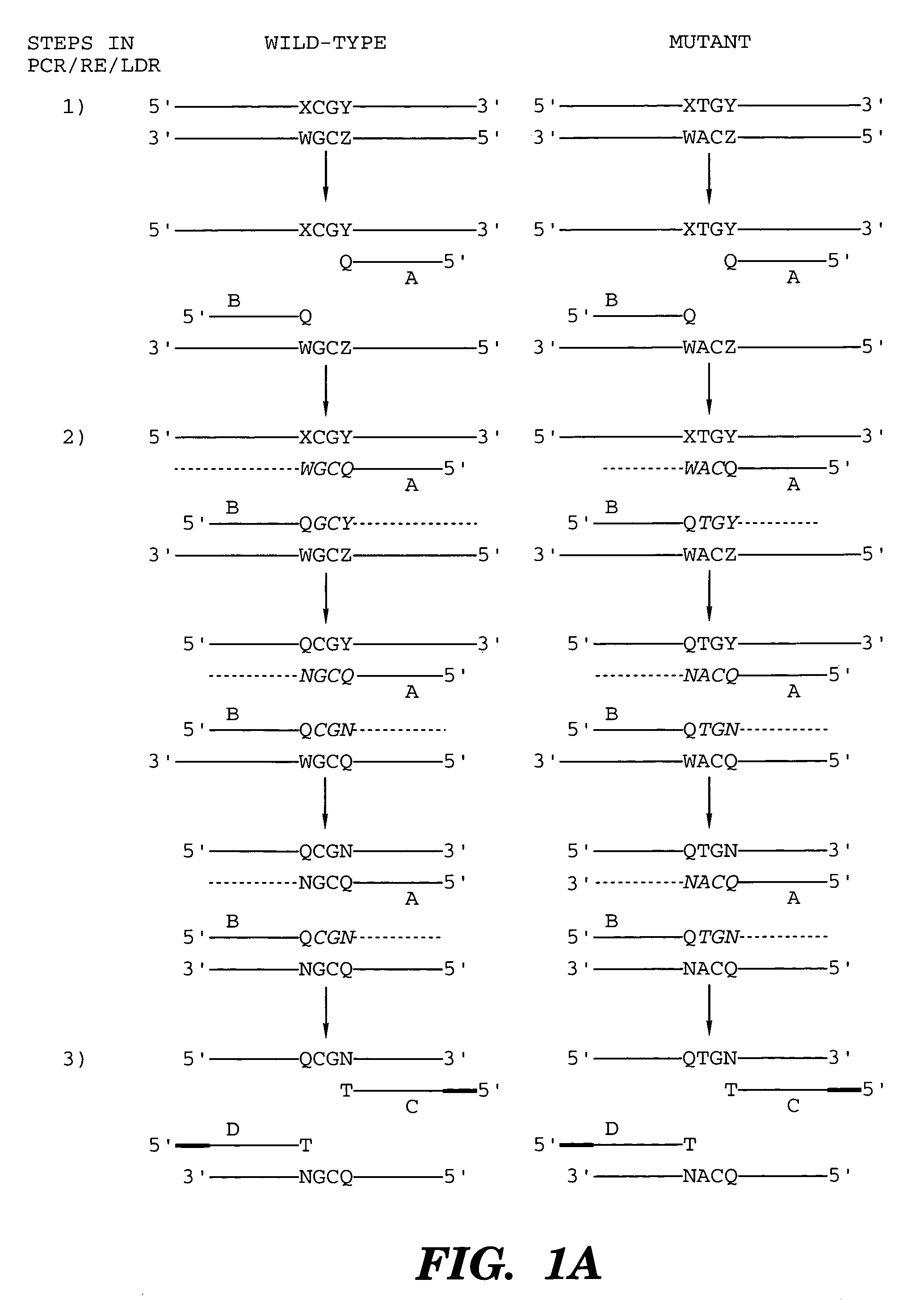
Coupled polymerase chain reaction-restriction-endonuclease digestion-ligase detection reaction process
InactiveUS7014994B1Sensitive highOptimizationSugar derivativesMicrobiological testing/measurementNucleotideWild type
The present invention provides a method for identifying one or more low abundance sequences differing by one or more single-base changes, insertions, or deletions, from a high abundance sequence in a plurality of target nucleotide sequences. The high abundance wild-type sequence is selectively removed using high fidelity polymerase chain reaction analog conversion, facilitated by optimal buffer conditions, to create a restriction endonuclease site in the high abundance wild-type gene, but not in the low abundance mutant gene. This allows for digestion of the high abundance DNA. Subsequently the low abundant mutant DNA is amplified and detected by the ligase detection reaction assay. The present invention also relates to a kit for carrying out this procedure.
Owner:LOUISIANA STATE UNIV +1
Promoter and plasmid system for genetic engineering
This invention provides a series of low-copy number plasmids comprising restriction endonuclease recognition sites useful for cloning at least three different genes or operons, each flanked by a terminator sequence, the plasmids containing variants of glucose isomerase promoters for varying levels of protein expression. The materials and methods are useful for genetic engineering in microorganisms, especially where multiple genetic insertions are sought.
Owner:EI DU PONT DE NEMOURS & CO
Determination of methylated DNA
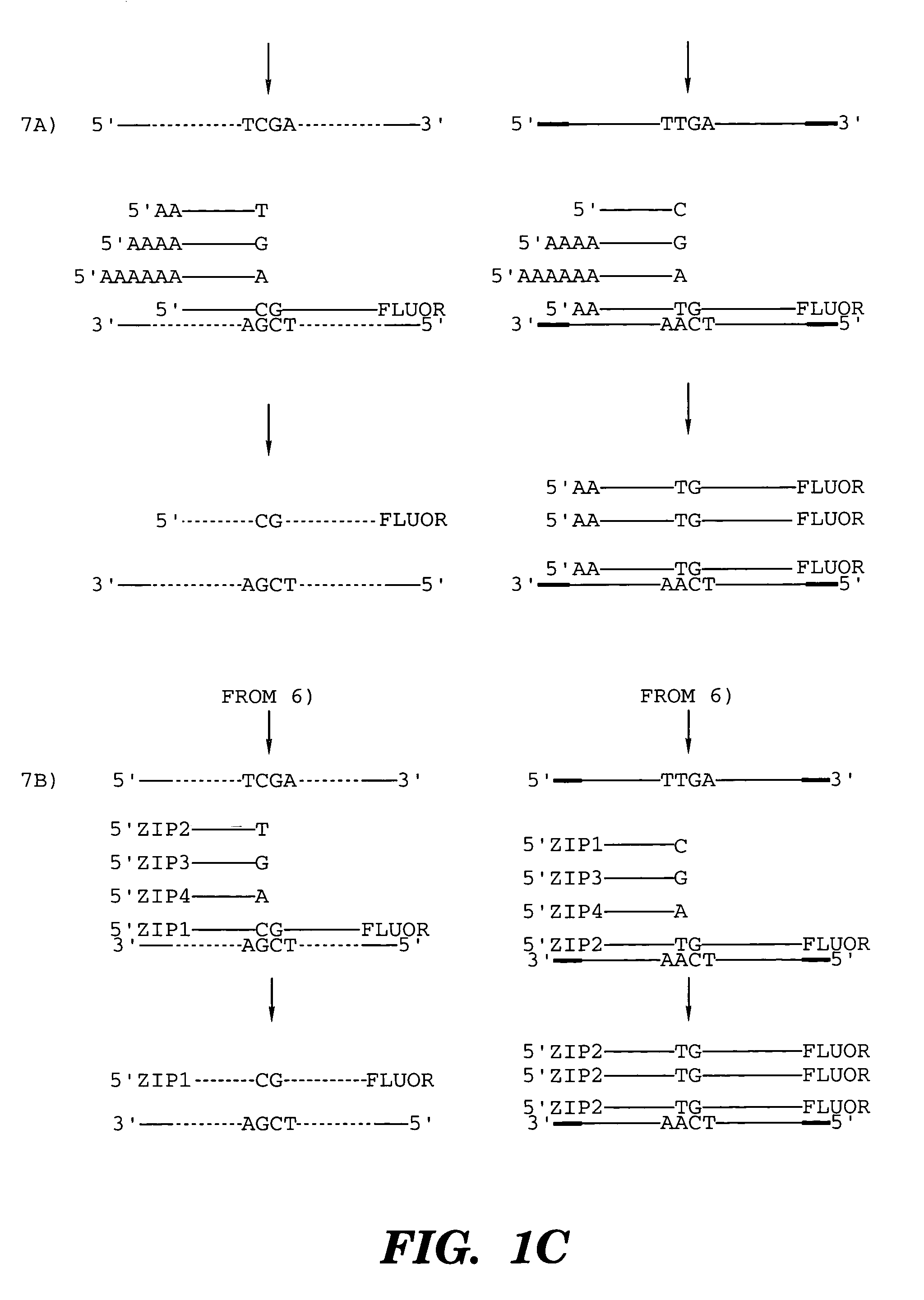
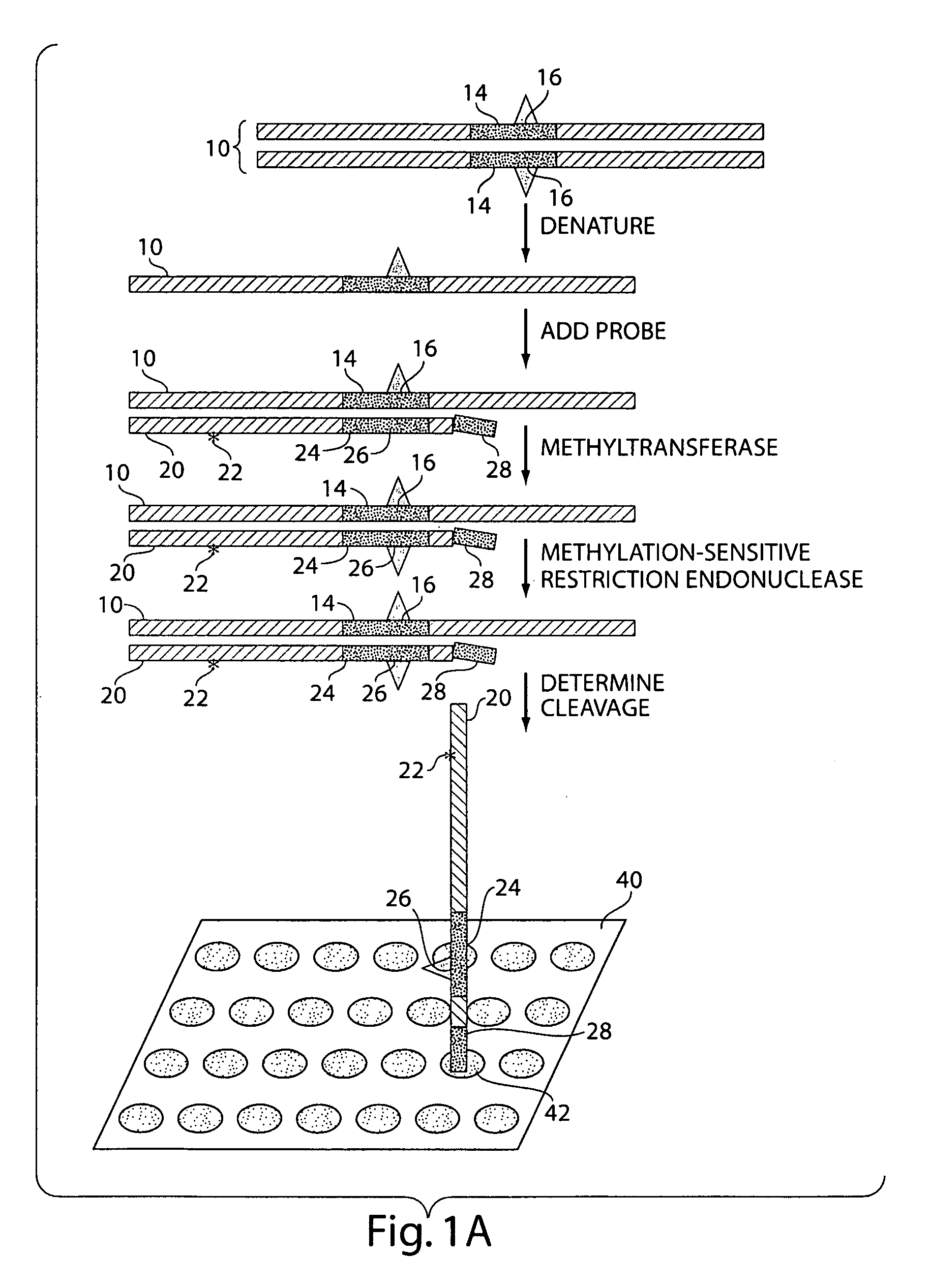
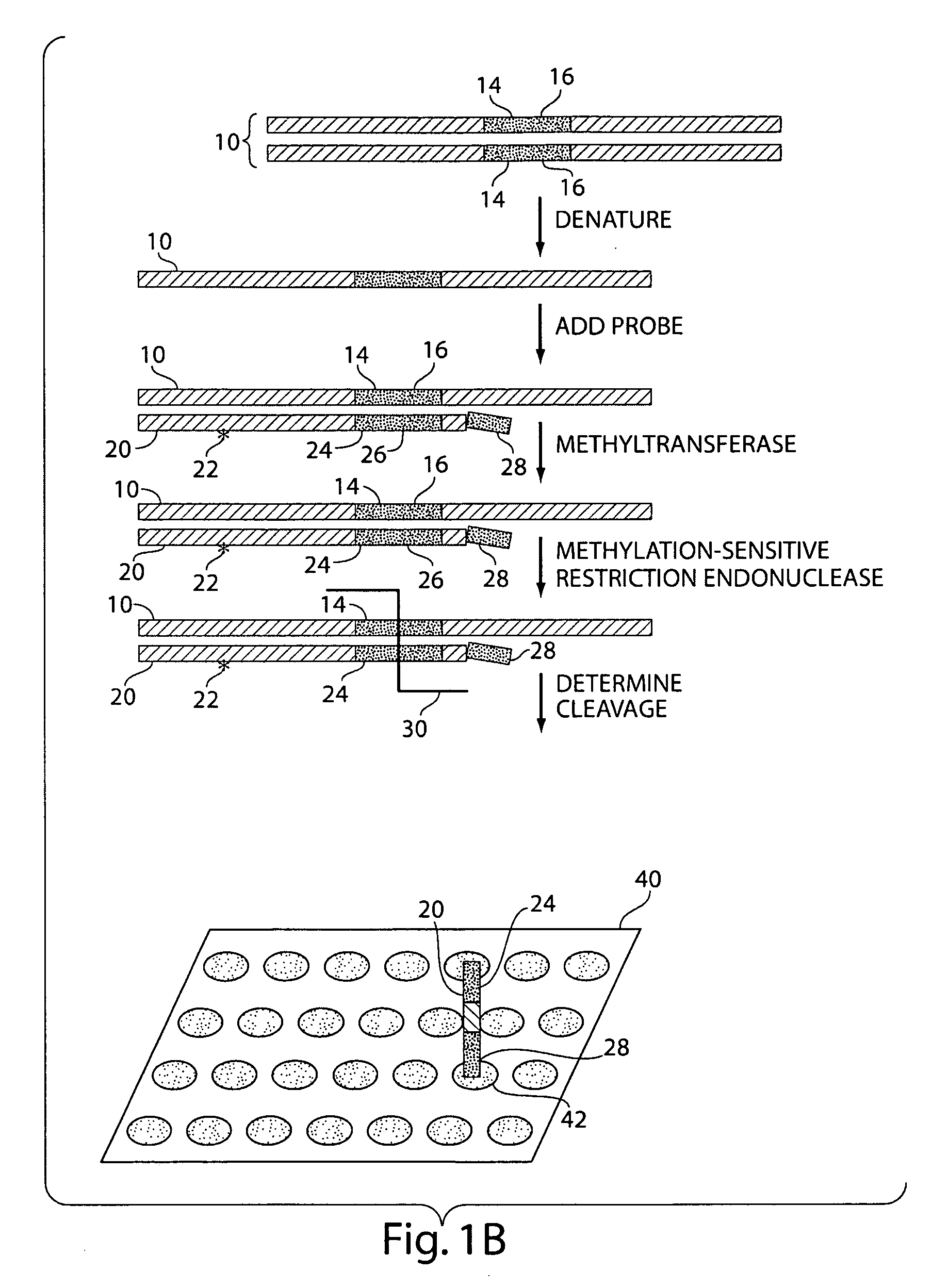
InactiveUS20070231800A1Microbiological testing/measurementFermentationNucleic Acid ProbesNucleic acid hybridisation
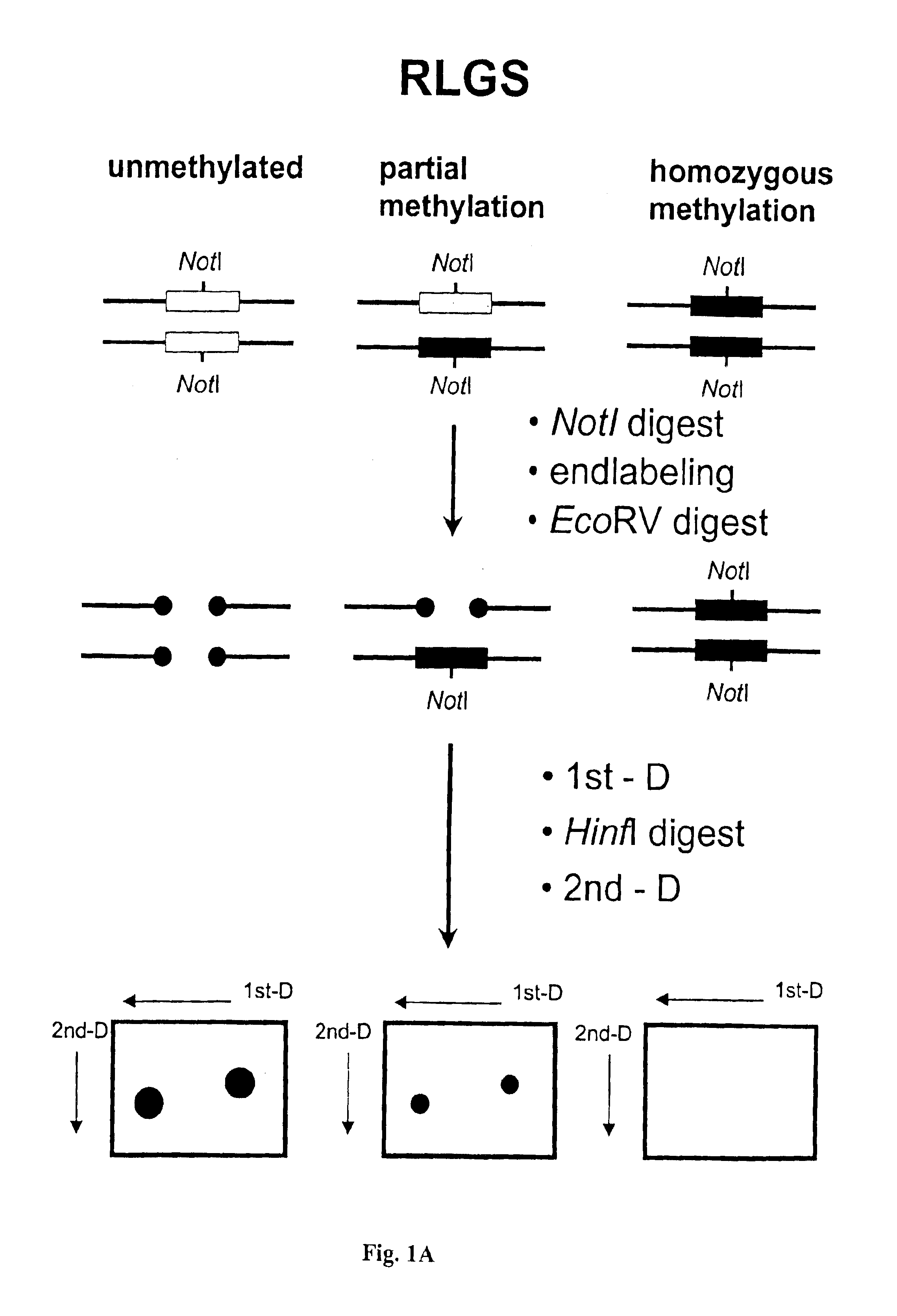
The present invention generally relates to the determination of the state of one or more locations within a nucleic acid and, in particular, to the determination of the methylation state of one or more methylation sites within a nucleic acid such as DNA. In one aspect of the invention, a nucleic acid, such as DNA, that is suspected of being methylated is exposed to a nucleic acid probe able to hybridize the nucleic acid at or near the methylation site. After hybridization, the nucleic acid-probe hybrid is exposed to a methylation-sensitive restriction endonuclease able to bind at or near the methylation site. The restriction endonuclease is not able to cleave the nucleic acid-probe hybrid if the DNA is methylated at the methylation site, but is able to cleave the nucleic acid-probe hybrid if the nucleic acid is not methylated at the methylation site. Determination of the cleavage state of the probe can thus be used to determine the state of the methylation site.
Owner:AGILENT TECH INC
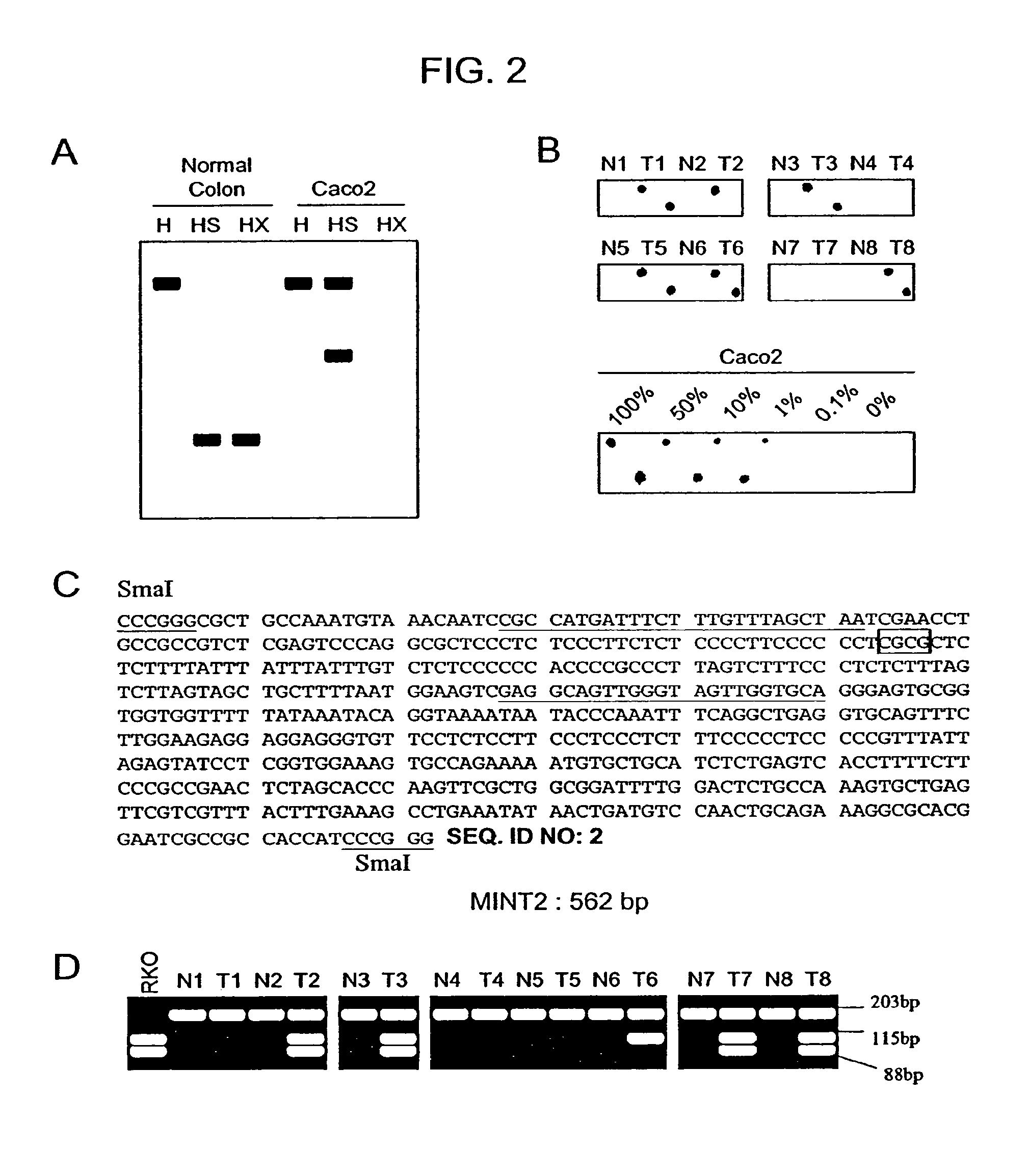
Detection of methylated CpG rich sequences diagnostic for malignant cells
InactiveUS6893820B1Microbiological testing/measurementMaterial analysis by electric/magnetic meansAbnormal tissue growthCancer cell
The present invention provides methods for determining the methylation status of CpG-containing dinucleotides on a genome-wide scale using infrequent cleaving, methylation sensitive restriction endonucleases and two-dimensional gel electrophoretic display of the resulting DNA fragments. Such methods can be used to diagnose cancer, classify tumors and provide prognoses for cancer patients. The present invention also provides isolated polynucleotides and oligonucleotides comprising CpG dinucleotides that are differentially methylated in malignant cells as compared to normal, non-malignant cells. Such polynucleotides and oligonucleotides are useful for diagnosis of cancer. The present invention also provides methods for identifying new DNA clones within a library that contain specific CpG dinucleotides that are differentially methylated in cancer cells as compared to normal cells.
Owner:THE OHIO STATE UNIV RES FOUND
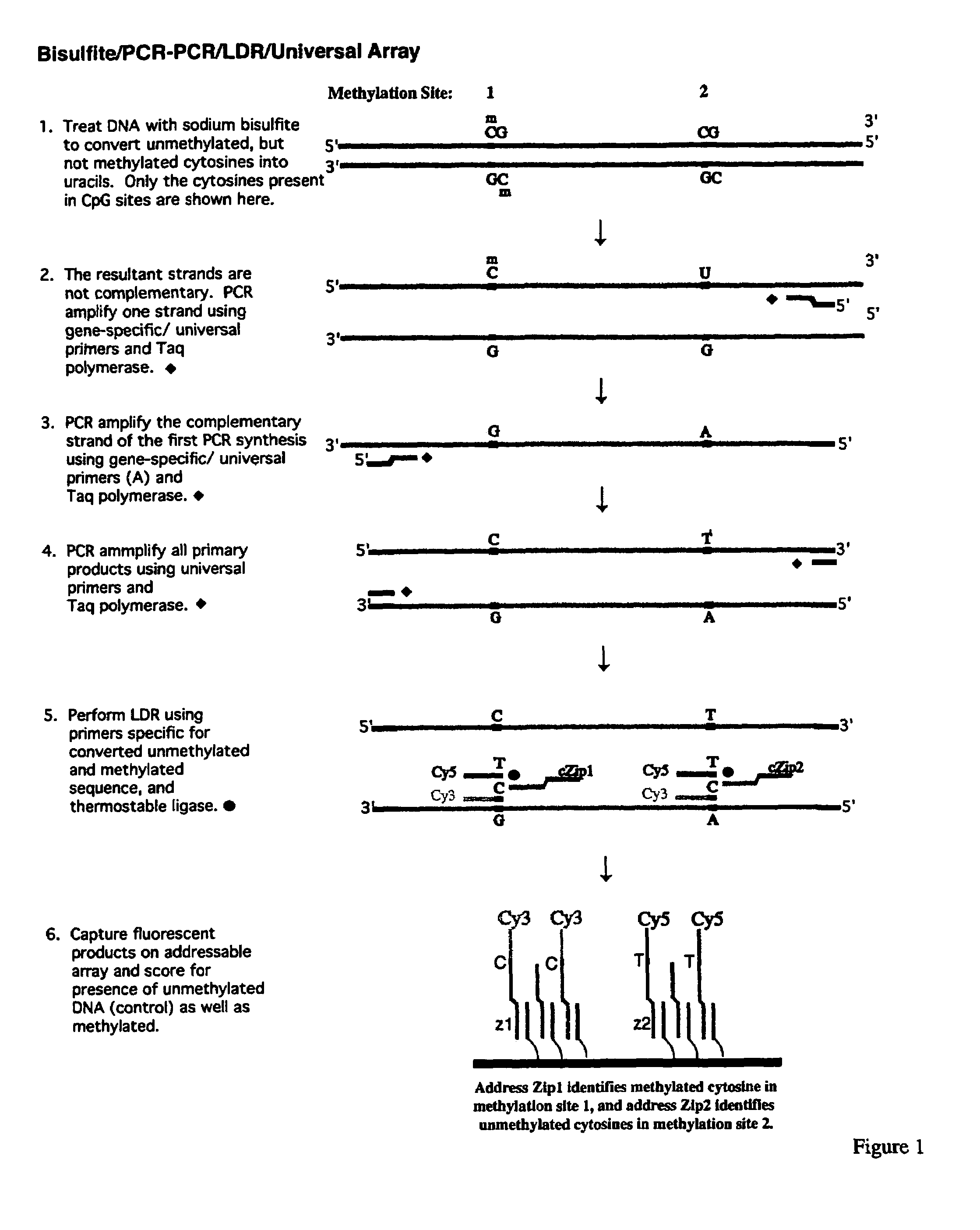
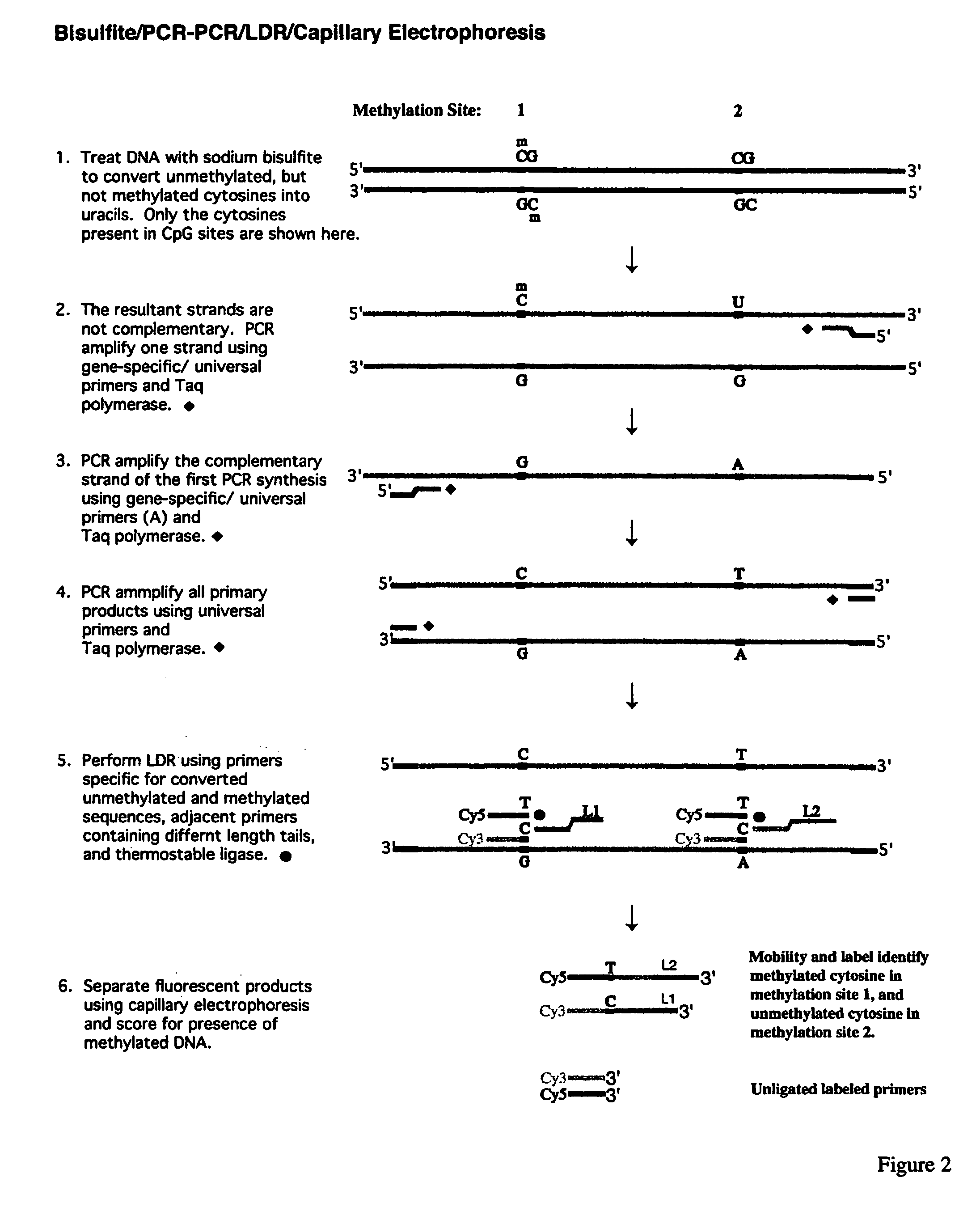
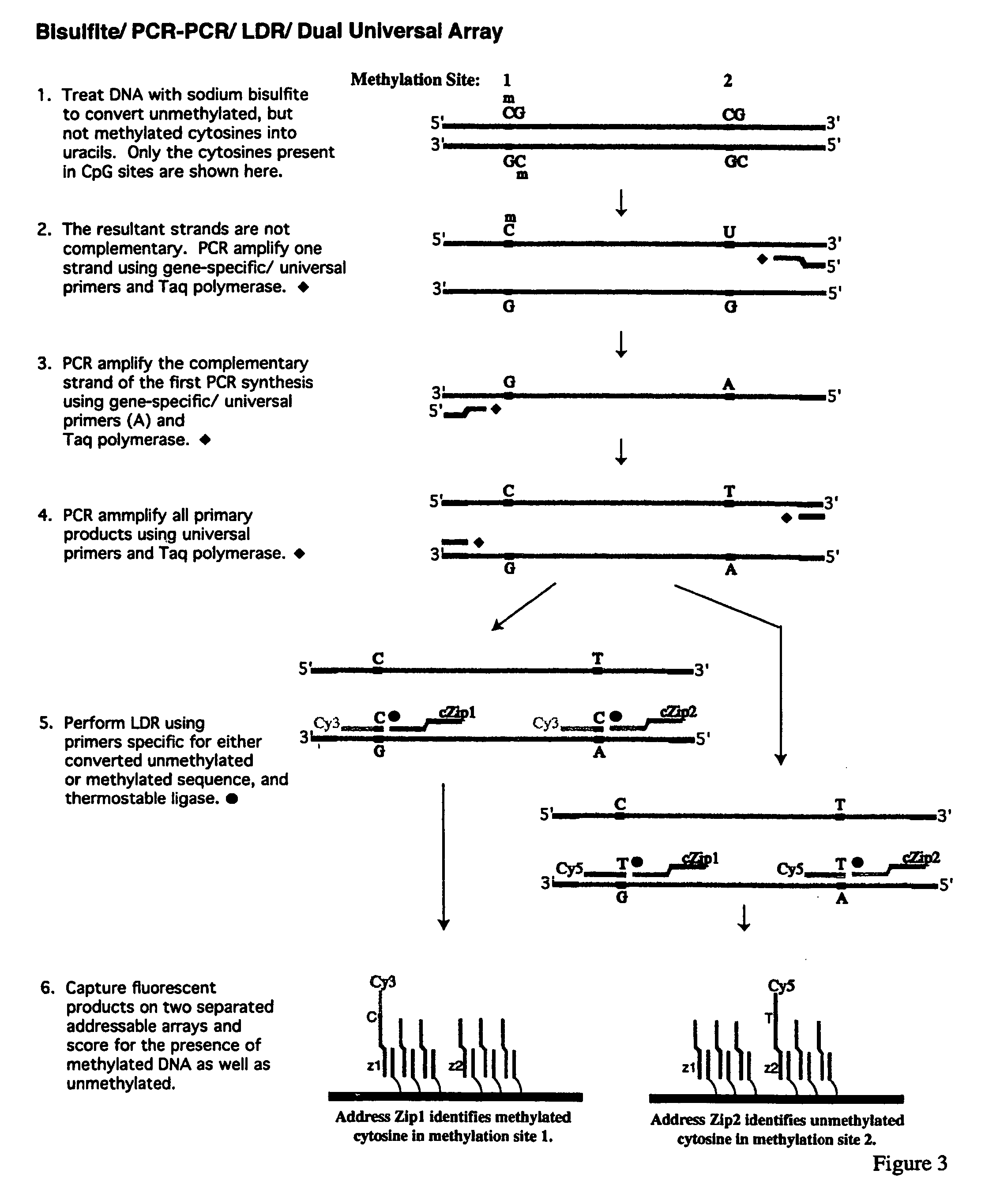
Method for detection of promoter methylation status
InactiveUS7358048B2Precise “ molecular signature ”Accurately methylation statusMicrobiological testing/measurementFermentationCapillary electrophoresisElectrophoresis
The present invention relates to the detection of promoter methylation status using a combination of either modification of methylated DNA or restriction endonuclease digestion, multiplex polymerase chain reaction, ligase detection reaction, and a universal array or capillary electrophoresis detection.
Owner:CORNELL RES FOUNDATION INC
Method and apparatus for identifying, classifying, or quantifying DNA sequences in a sample without sequencing
InactiveUS6418382B2Rapid and economical and quantitative and precise determination and classificationSufficient discrimination and resolutionData processing applicationsMicrobiological testing/measurementSample sequenceSingle sequence
This invention provides methods by which biologically derived DNA sequences in a mixed sample or in an arrayed single sequence clone can be determined and classified without sequencing. The methods make use of information on the presence of carefully chosen target subsequences, typically of length from 4 to 8 base pairs, and preferably the length between target subsequences in a sample DNA sequence together with DNA sequence databases containing lists of sequences likely to be present in the sample to determine a sample sequence. The preferred method uses restriction endonucleases to recognize target subsequences and cut the sample sequence. Then carefully chosen recognition moieties are ligated to the cut fragments, the fragments amplified, and the experimental observation made. Polymerase chain reaction (PCR) is the preferred method of amplification. Another embodiment of the invention uses information on the presence or absence of carefully chosen target subsequences in a single sequence clone together with DNA sequence databases to determine the clone sequence. Computer implemented methods are provided to analyze the experimental results and to determine the sample sequences in question and to carefully choose target subsequences in order that experiments yield a maximum amount of information.
Owner:CURAGEN CORP
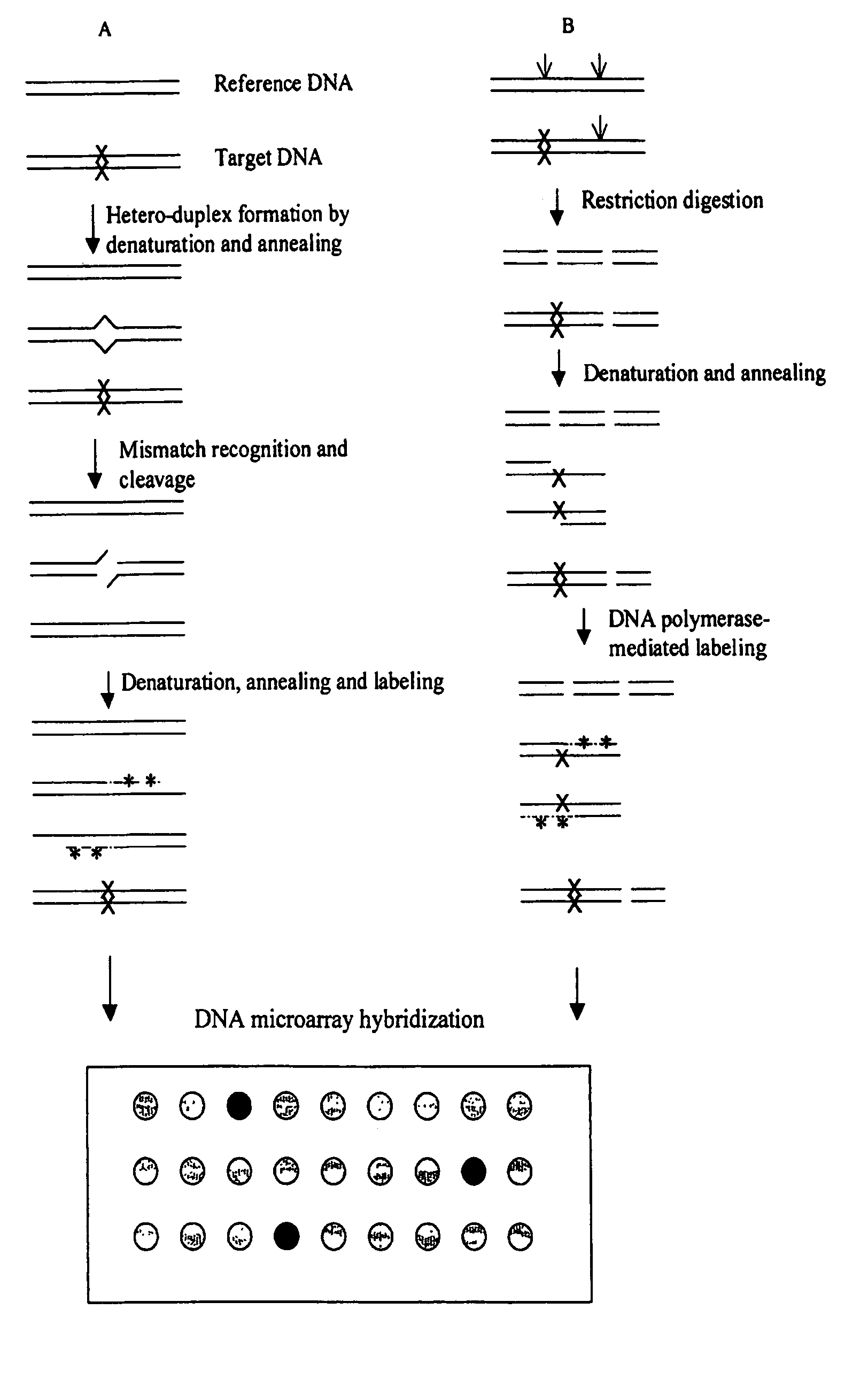
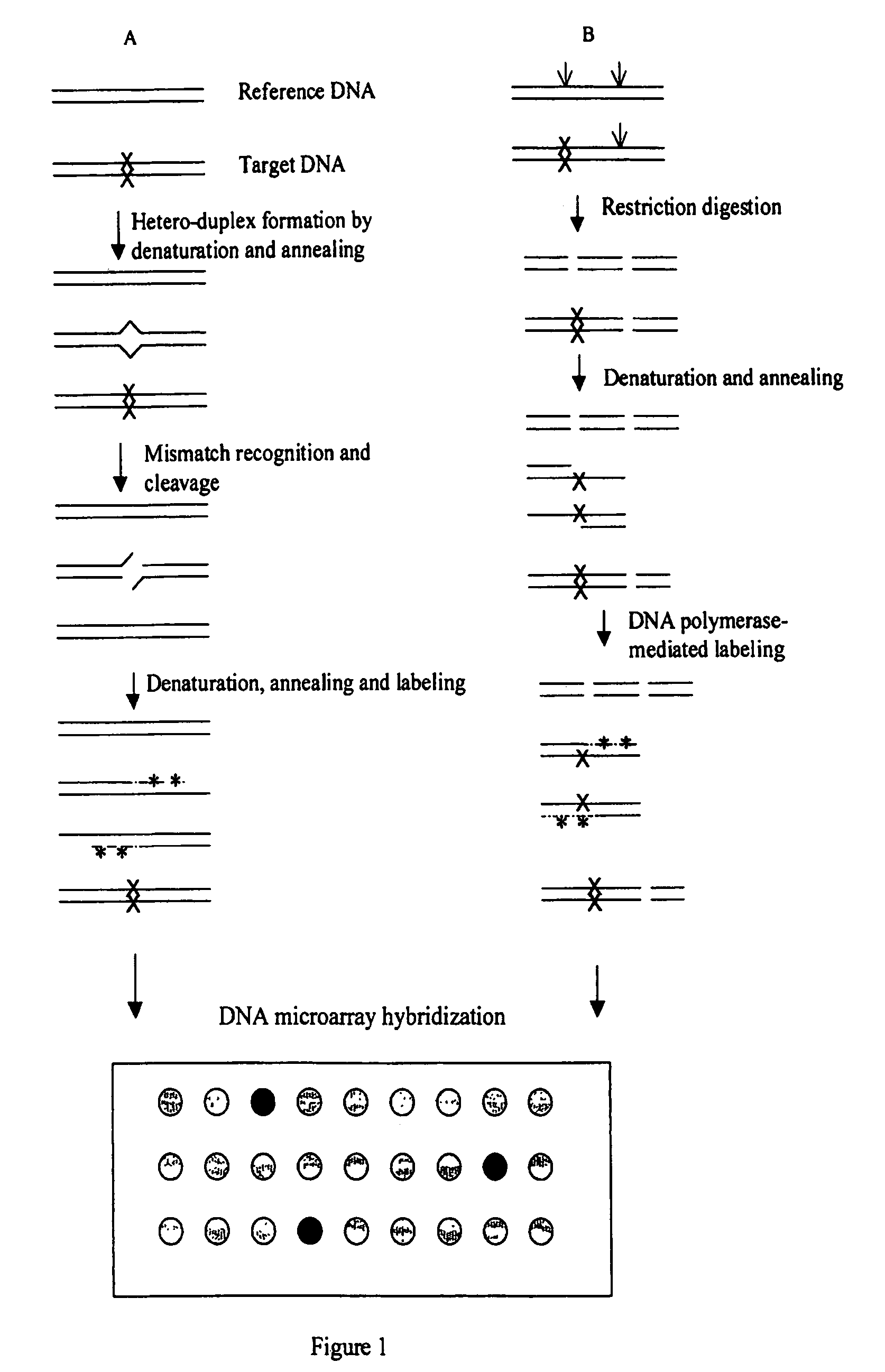
Methods for detecting and localizing DNA mutations by microarray
InactiveUS7378245B2Reduce impactMicrobiological testing/measurementFermentationDNA microarrayHeteroduplex
Owner:UNIVERSITY OF OREGON
Method to clone mRNAs
InactiveUS6261770B1Convenient verificationHigh homologySugar derivativesMicrobiological testing/measurementNucleotideNucleotide sequencing
Disclosed and claimed is a method for preparing a normalized sub-divided library of amplified cDNA fragments from the coding region of mRNAs contained in a sample. The method includes the steps of: a) subjecting the mRNA population to reverse transcription using at least one cDNA primer, thereby obtaining first strand cDNA fragments, b) synthesizing second strand cDNA complementary to the first strand cDNA fragments by use of the first strand DNA fragments as templates, thereby obtaining double stranded cDNA fragments, c) digesting the double stranded cDNA fragments with at least one restriction endonuclease, the endonuclease leaving protruding sticky ends of similar size at the termini of the DNA after digestion, thereby obtaining cleaved cDNA fragments, d) adding at least two adapter fragments containing known sequences to the cleaved cDNA fragments obtained in step c), the at least two adapter fragments being able to bind specifically to the sticky ends of the double stranded cDNA produced in step c), the one adapter fragment being able to anneal to the primer having formula I in step f), the second adapter fragment being a termination fragment introducing a block against DNA polymerization in the 5'->3' direction setting out from said termination fragment and the termination fragment being unable to anneal to any primer of the at least two primer sets in step f) during the molecular amplification procedure, the at least two adapter fragments being ligated to the cleaved cDNA fragments obtained in step c) so as to obtain ligated cDNA fragments, e) sub-dividing the ligated cDNA fragments obtained in step d) into 4n1 pools where 1<=n1<=4, and f) subjecting each pool of ligated cDNA fragments obtained in step e) to a molecular amplification procedure so as to obtain amplified cDNA fragments, wherein is used, for an adapter fragment used in step d), a set of amplification primers having the general formula Iwherein Com is a sequence complementary to at least the 5'-end of an adapter fragment which is ligated to the 3'-end of a cleaved cDNA fragment, N is A, G, T, or C, the one primer having the general formula I where n1=0, and the second primer having the general formula I where 1<=n1<=4, the second primer being capable of priming amplification of any nucleotide sequence ligated in its 3'-end to the adapter fragment complementary in its 5'-end to Com.
Owner:AZIGN BIOSCI
Restriction endonucleases and their applications
Provided is a methylation-specific restriction endonuclease for a DNA duplex substrate, which endonuclease recognizes in a strand of the duplex a 2 to 6 nucleotide recognition sequence comprising a 5-methylcytosine, and cleaves each strand of the duplex at a fixed position outside the recognition sequence.
Owner:THERMO FISHER SCI BALTICS UAB
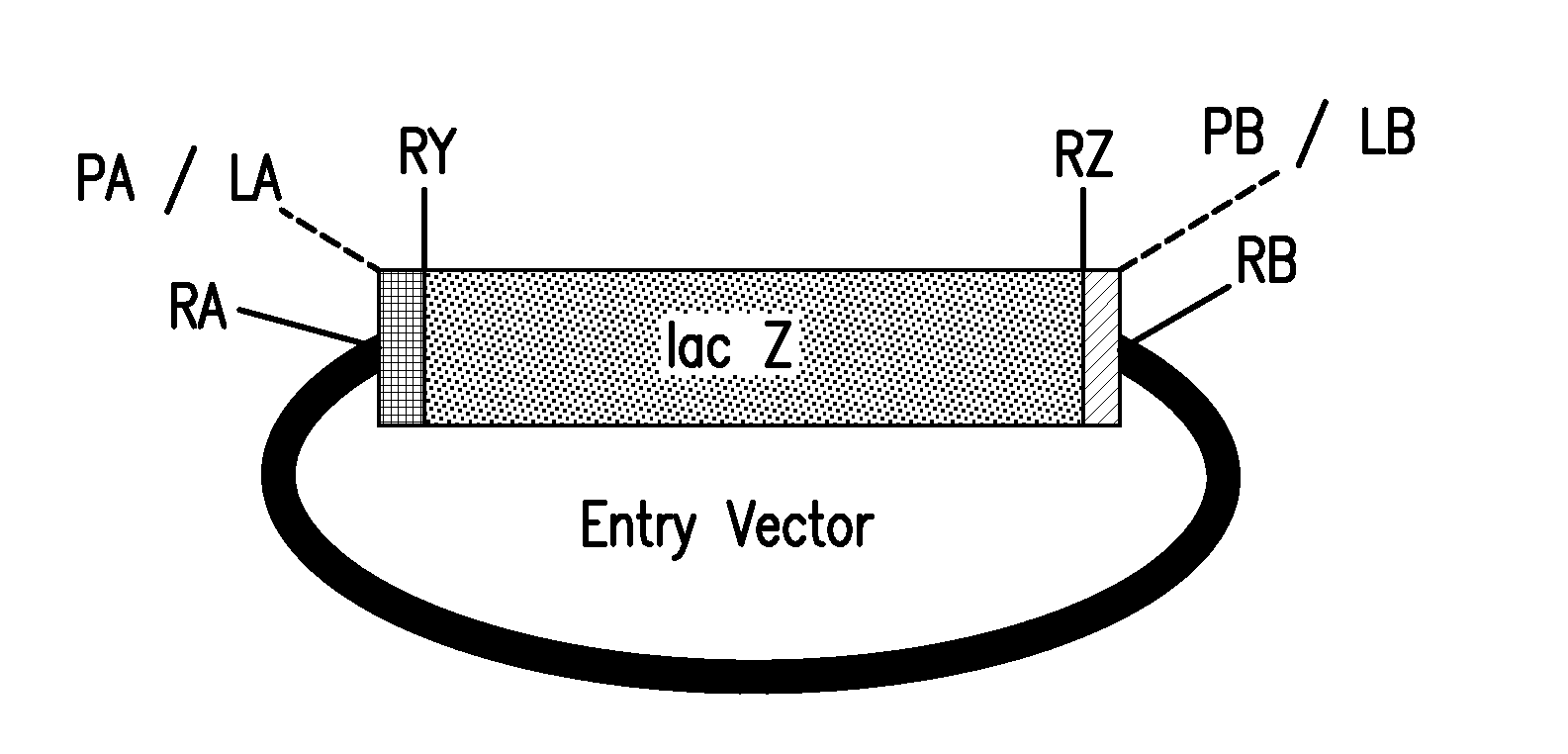
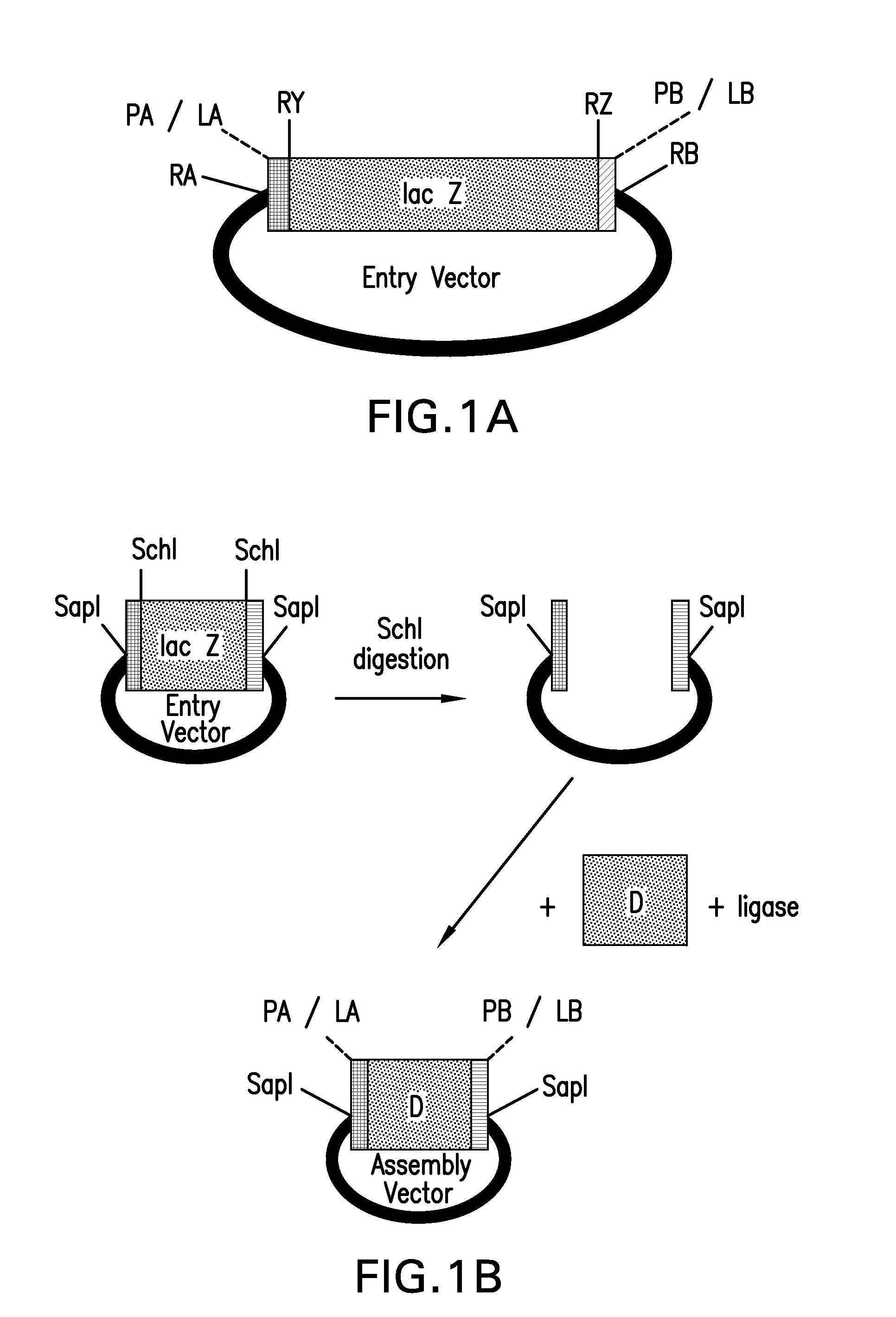
Method of cloning at least one nucleic acid molecule of interest using type iis restriction endonucleases, and corresponding cloning vectors, kits and system using type iis restriction endonucleases
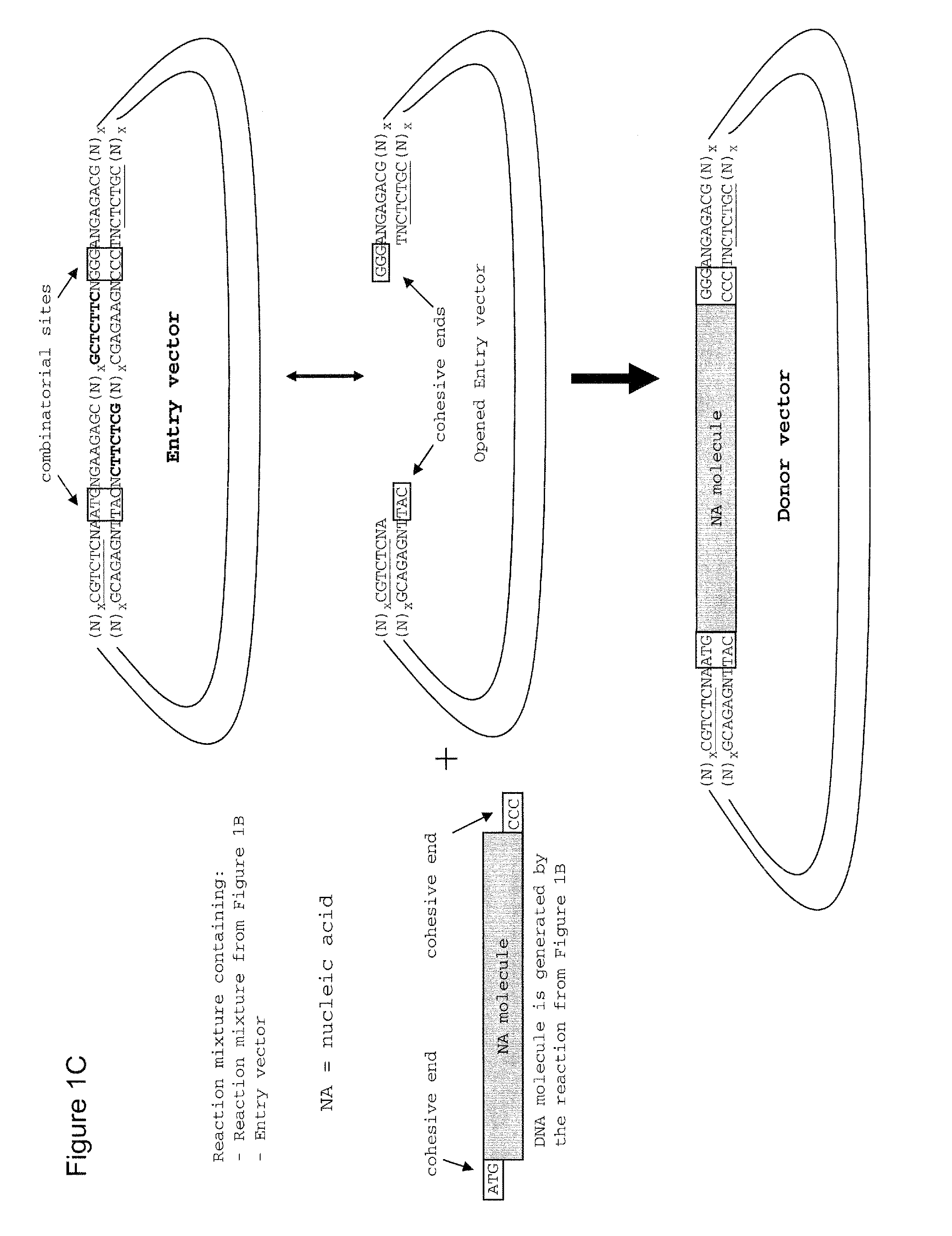
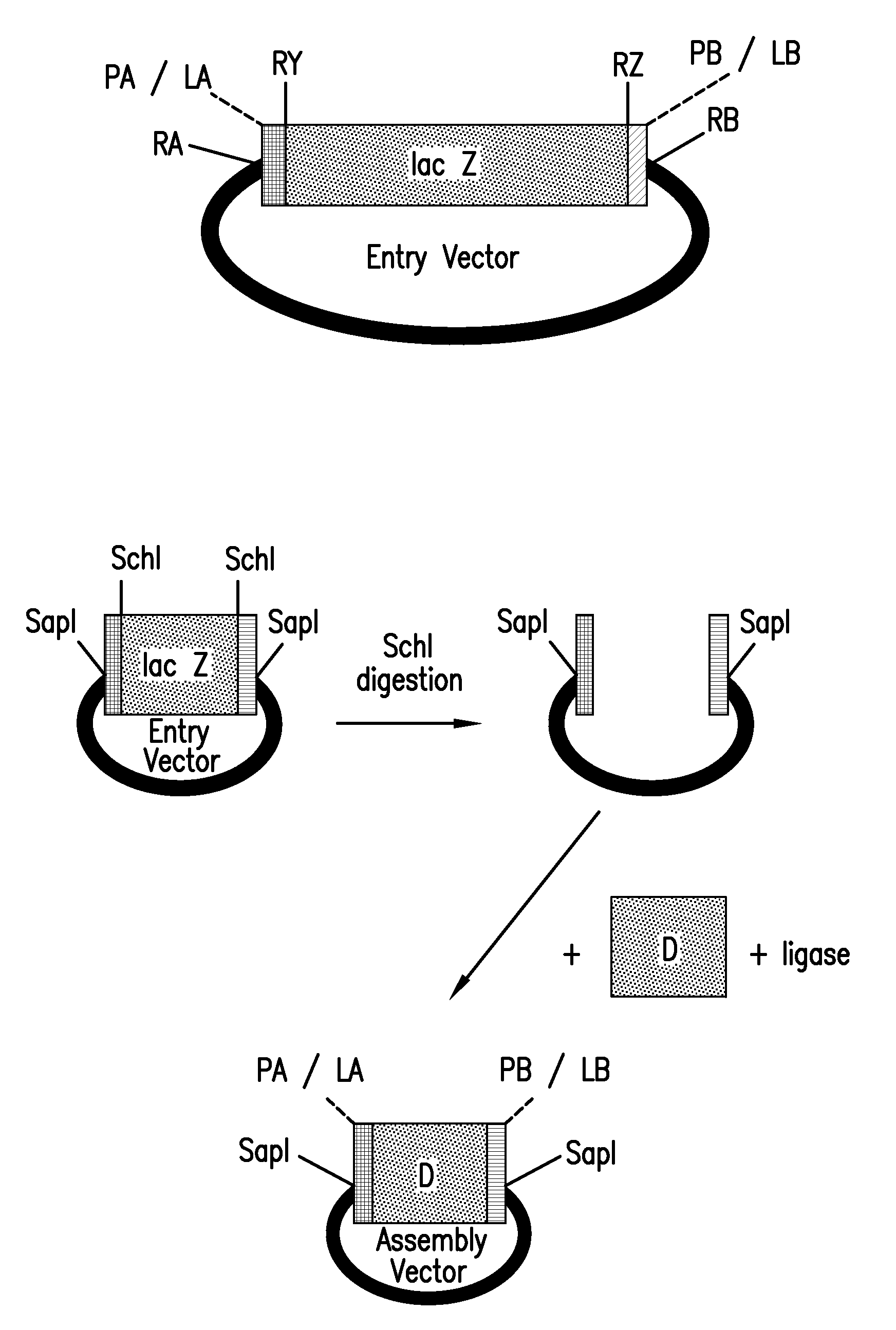
The present invention refers to methods of (sub)cloning at least one nucleic acid molecule of interest. One embodiment relates to a method of (sub)cloning at least one nucleic acid molecule of interest comprising a) providing at least one (replicable) Entry vector into which the at least one nucleic acid molecule of interest is to be inserted, wherein the at least one Entry vector carries two recognition sites for at least one first type IIS and / or type IIS like restriction endonuclease and wherein said at least one nucleic acid molecule of interest can be excised from the at least one Entry vector at two combinatorial sites with one (same) or more (different) cohesive ends that are formed by the at least one first type IIS or type IIS like restriction endonuclease, and b) providing an Acceptor vector, into which the at least one nucleic acid molecule of interest is transferred from the at least one Entry vector carrying the at least one nucleic acid molecule of interest, wherein said Acceptor vector comprises at least one recognition site for at least one second type IIS restriction endonuclease and / or at least one recognition sites for at least one type IIS like restriction endonuclease, and wherein said Acceptor vector provides two combinatorial sites identical to the two combinatorial sites present in the Entry vector. The inventions also relates respective cloning vector and kits.
Owner:PHILIPPS UNIV MARBURG
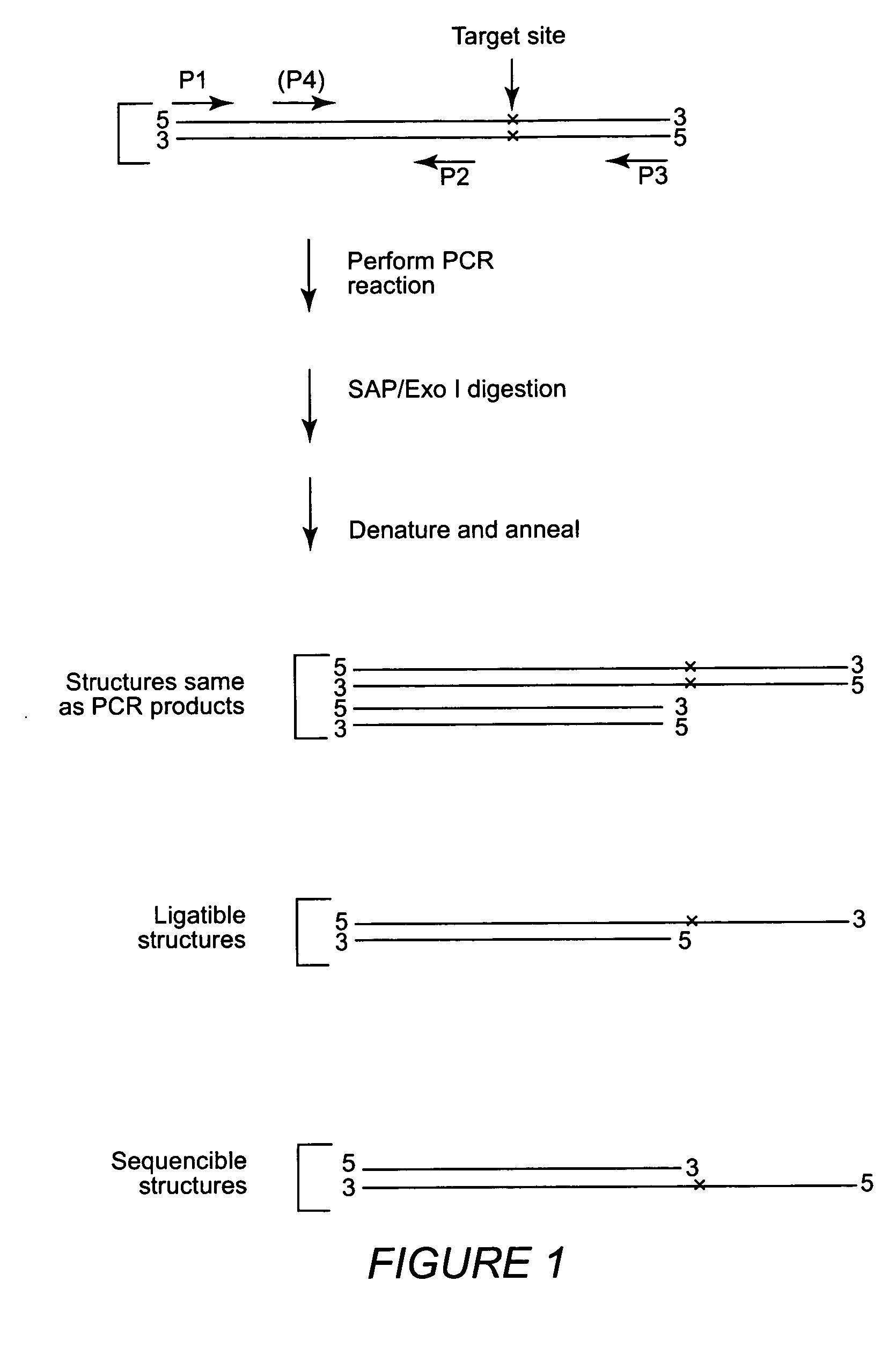
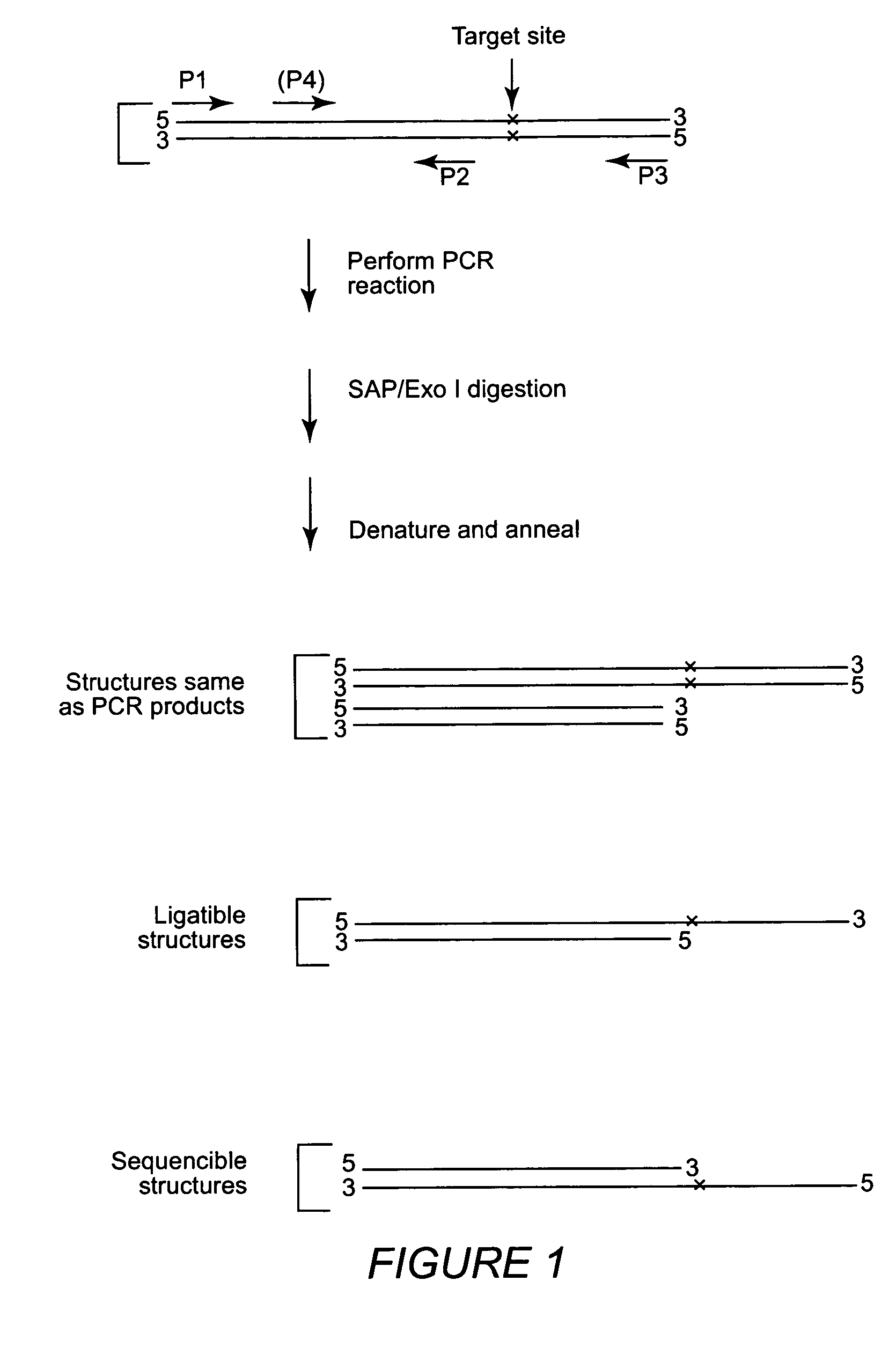
Multiple sequencible and ligatible structures for genomic analysis
InactiveUS20050175996A1Highly cost-effectiveImprove throughputMicrobiological testing/measurementFermentationNon targetedGenome
High throughput methods and kits for single nucleotide polymorphism (SNP) genotyping are provided. The methods involve utilizing nested PCR amplification reactions which produce sequencible and ligatible structures. An outer PCR primer set amplifies the SNP, and an inner PCR primer set amplifies a portion of the DNA amplified by the outer primer set, but does not amplify the SNP itself. The inner and outer primers may reaction include non-target common domain sequences, and the inner primer common domain sequences may comprise digestion restriction endonuclease recognition sites. The design of the inner primer set allows precise tailoring of the sequencible and ligatible structures with respect to length and base composition.
Owner:VIRGINIA COMMONWEALTH UNIV
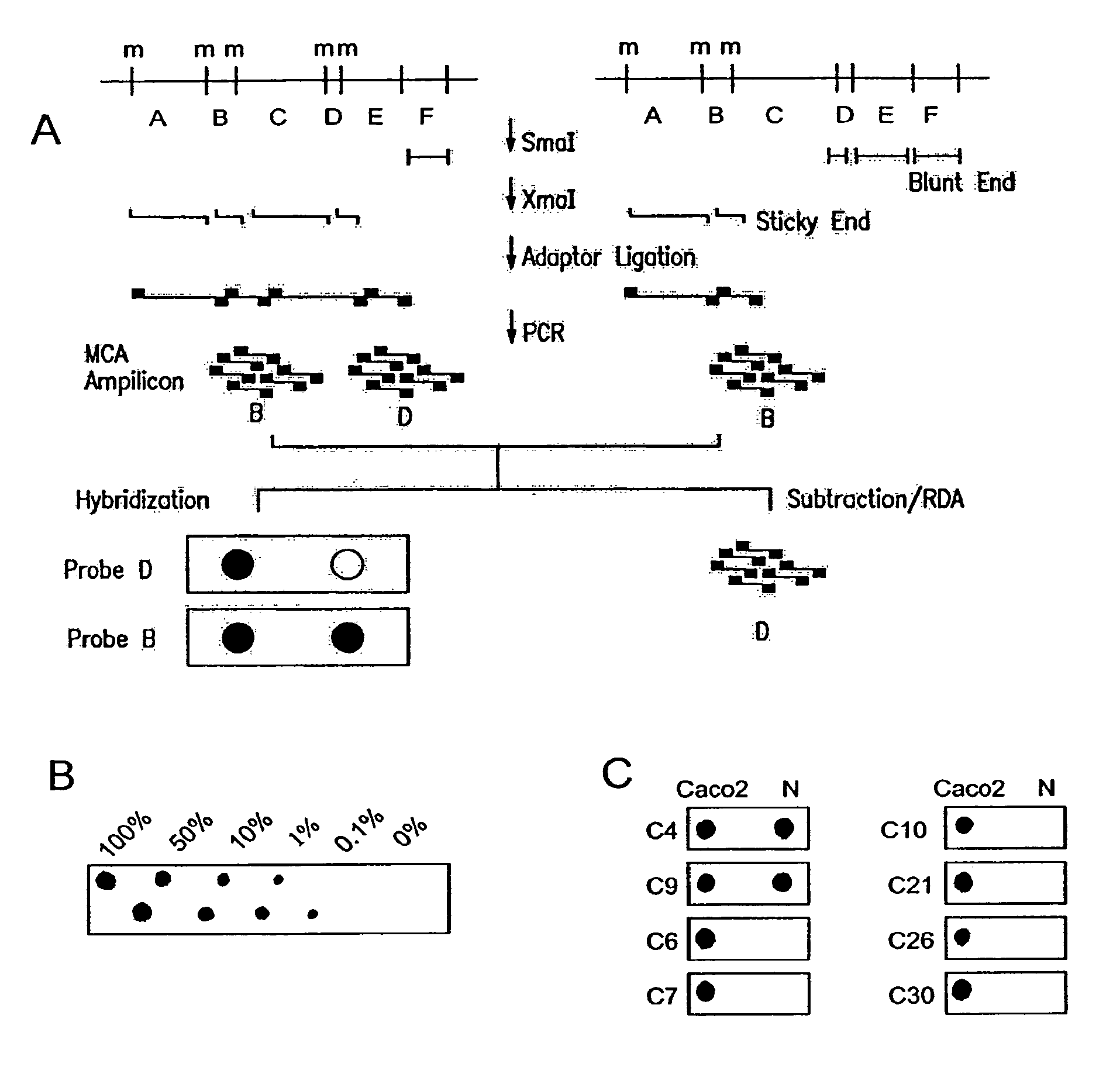
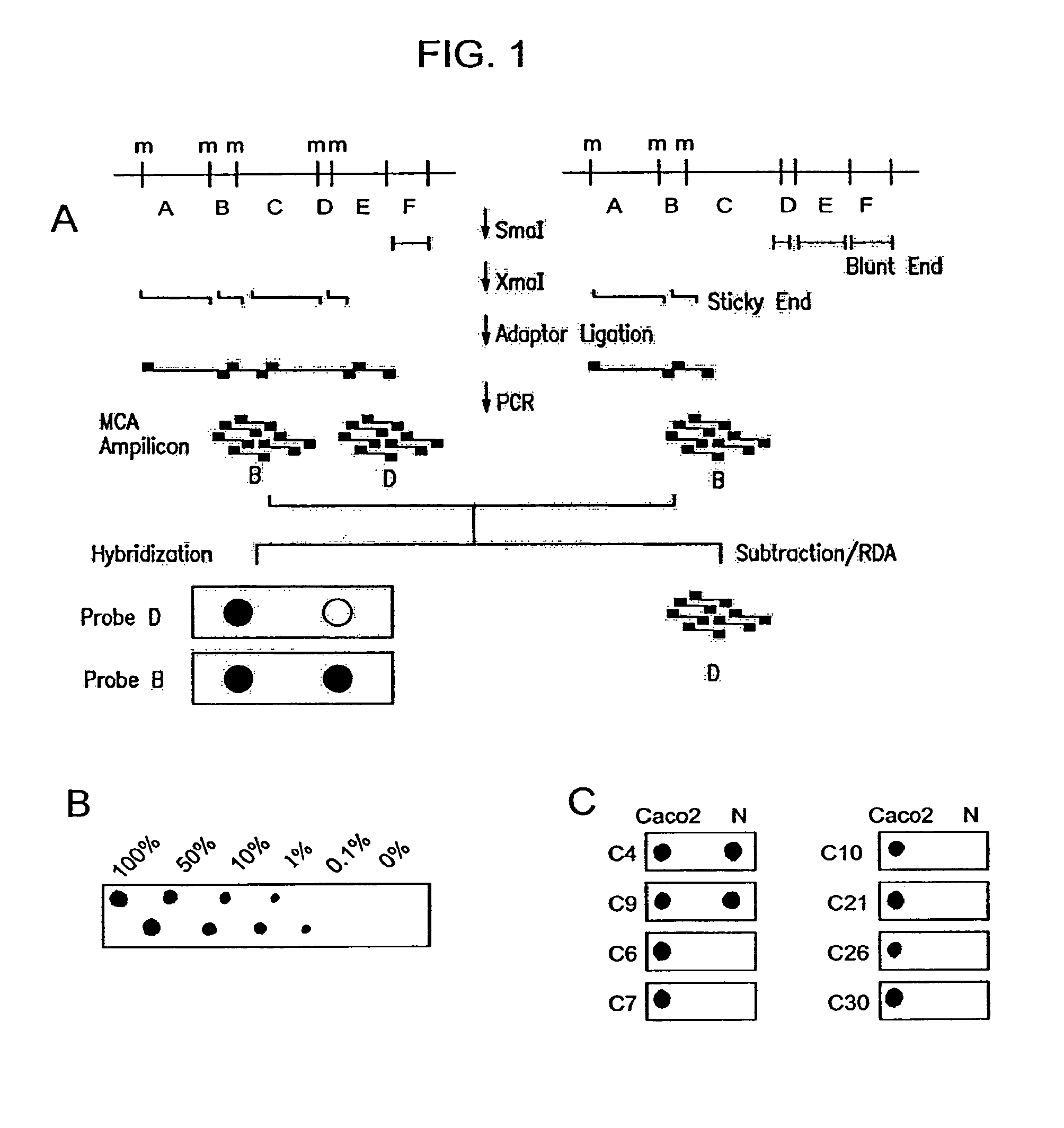
Methylated CpG island amplification (MCA)
The present invention provides a method for identifying a methylated CpG containing nucleic acid by contacting a nucleic acid with a methylation sensitive restriction endonuclease that cleaves unmethylated CpG sites and contacting the sample with an isoschizomer of the methylation sensitive restriction endonuclease, which cleaves both methylated and unmethylated CpG sites. The method also includes amplification of the CpG-containing nucleic acid using CpG-specific oligonucleotide primers A method is also provided for detecting an age associated disorder by identification of a methylated CpG containing nucleic acid. A method is further provided for evaluation the response of a cell to an agent A kit useful for detection of a CpG containing nucleic acid is also provided. Nucleic acid sequences encoding novel methylated clones.
Owner:THE JOHN HOPKINS UNIV SCHOOL OF MEDICINE
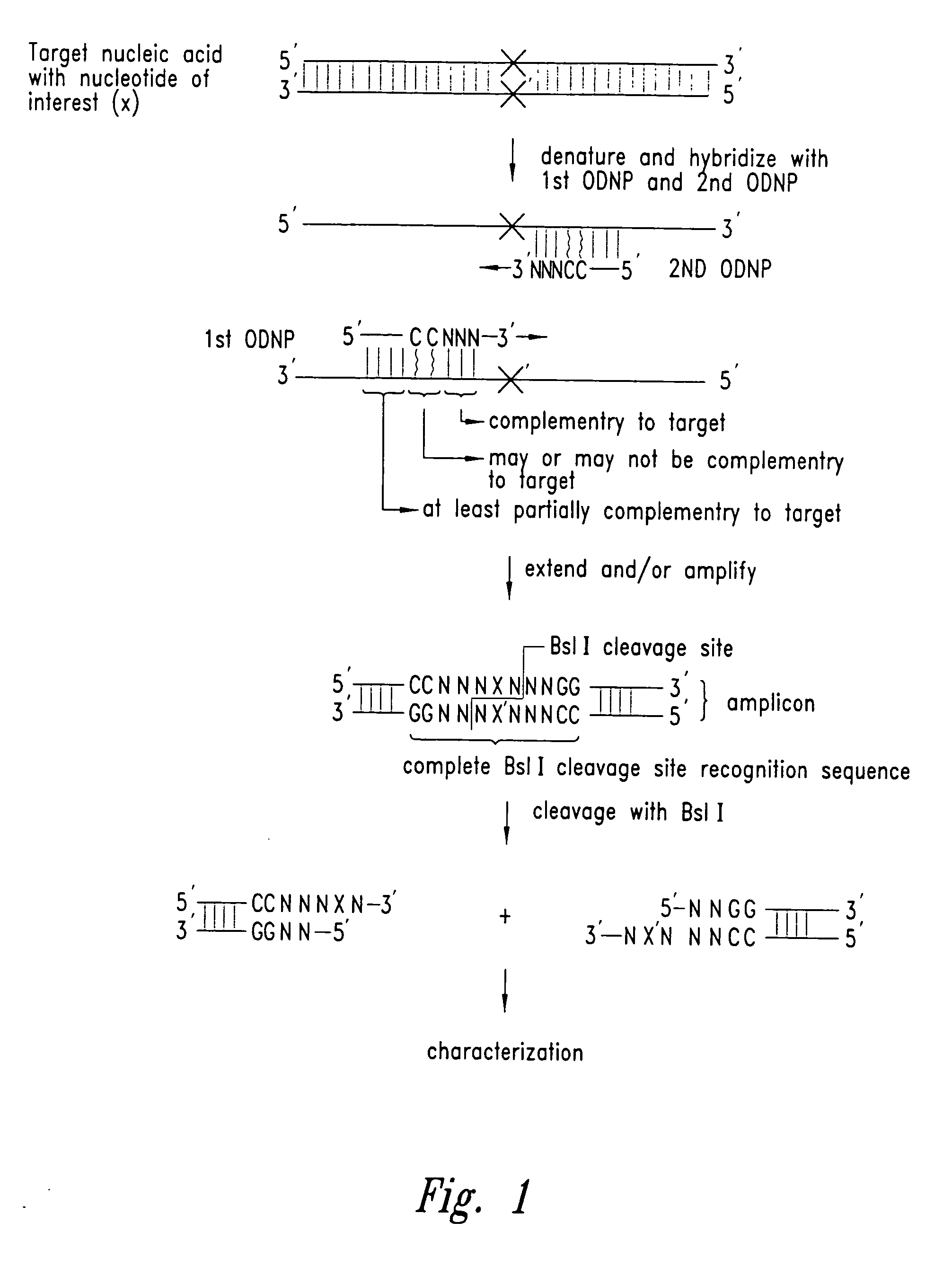
Methods for identifying nucleotides at defined positions in target nucleic acids
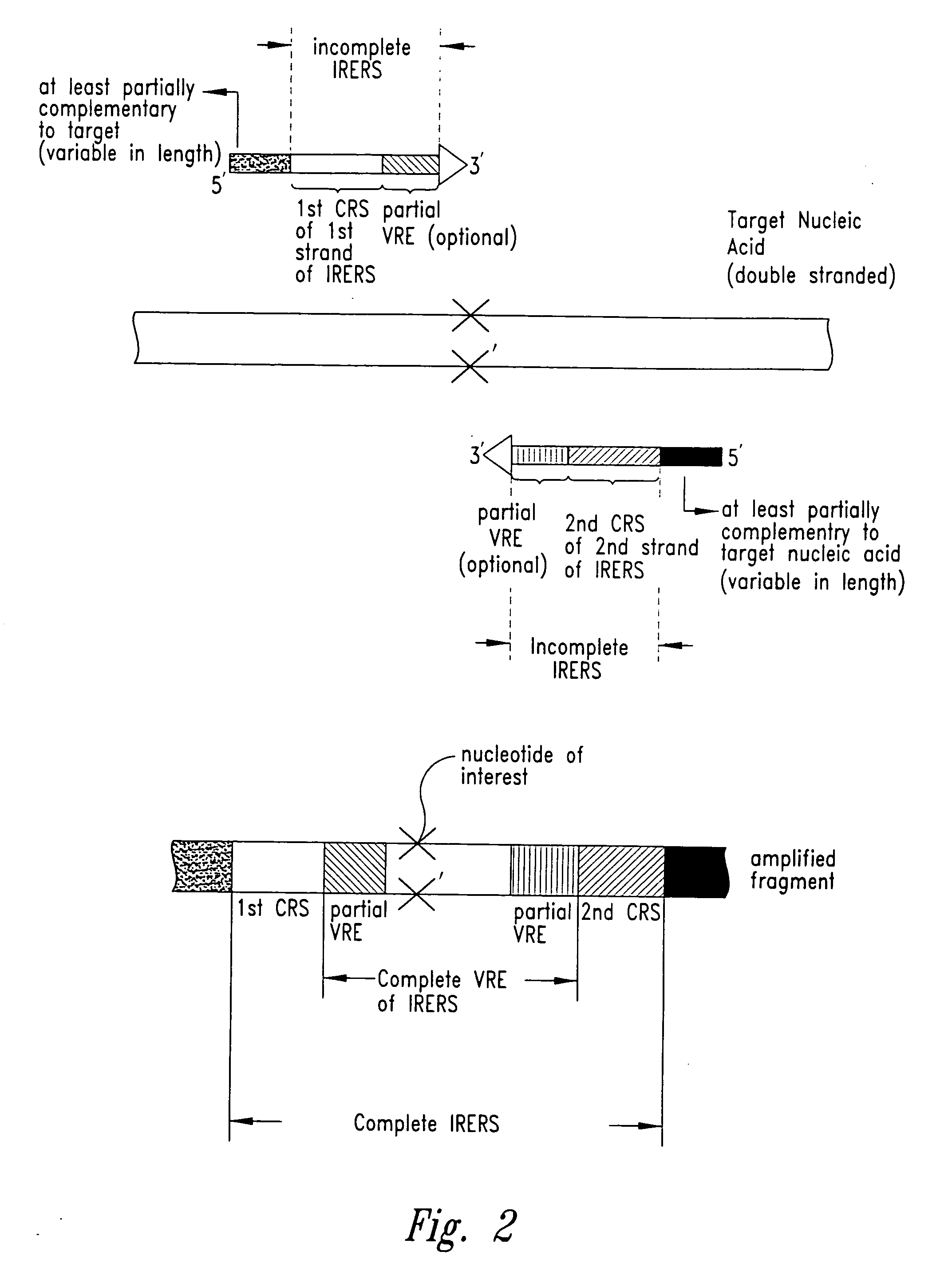
The identity of a nucleotide of interest in a target nucleic acid molecule is determined by combining the target with two primers, where the first primer hybridizes to and extends from a location 3' of the nucleotide of interest in the target, so as to incorporate the complement of the nucleotide of interest in a first extension product. The second primer then hybridizes to and extends based on the first extension product, at a location 3' of the complement of the nucleotide of interest, so as to incorporate the nucleotide of interest in a second extension product. The first primer then hybridizes to and extends from a location 3' of the nucleotide of interest in the second extension product, so as to form, in combination with the second extension product, a nucleic acid fragment. The first and second primers are designed to incorporate a portion of the recognition sequence of a restriction endonuclease that recognizes a partially variable interrupted base sequence. i.e. a sequence of the form A-B-C where A and C are a number and sequence of bases essential for RE recognition, and B is a number of bases essential for RE recognition. The first primer incorporates the sequence A, the second primer incorporates the sequence C, and they are designed, in view of the target, to product a nucleic acid fragment where sequences A and C are separated by the bases B, where the nucleotide of interest is within region B. Action of the RE on the nucleic acid fragment provides a small nucleic acid fragment that is amendable to characterization, to thereby reveal the identity of the nucleotide of interest.
Owner:KECK GRADUATE INST OF APPLIED LIFE SCI
Kits for amplifying DNA
ActiveUS8183359B2Reduce appearance problemsMinimize and substantially eliminate emergenceSugar derivativesMicrobiological testing/measurementAmplification dnaA-DNA
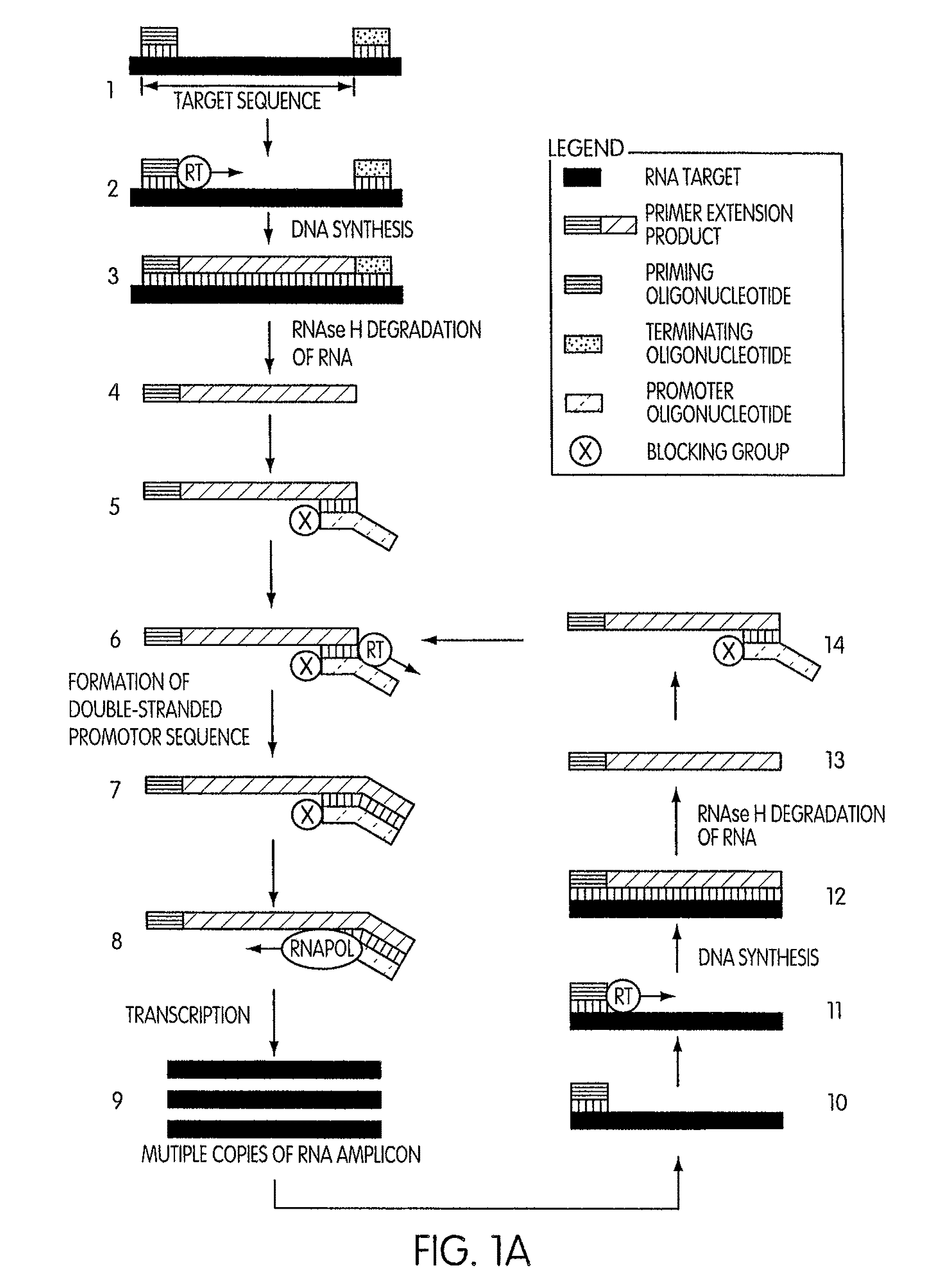
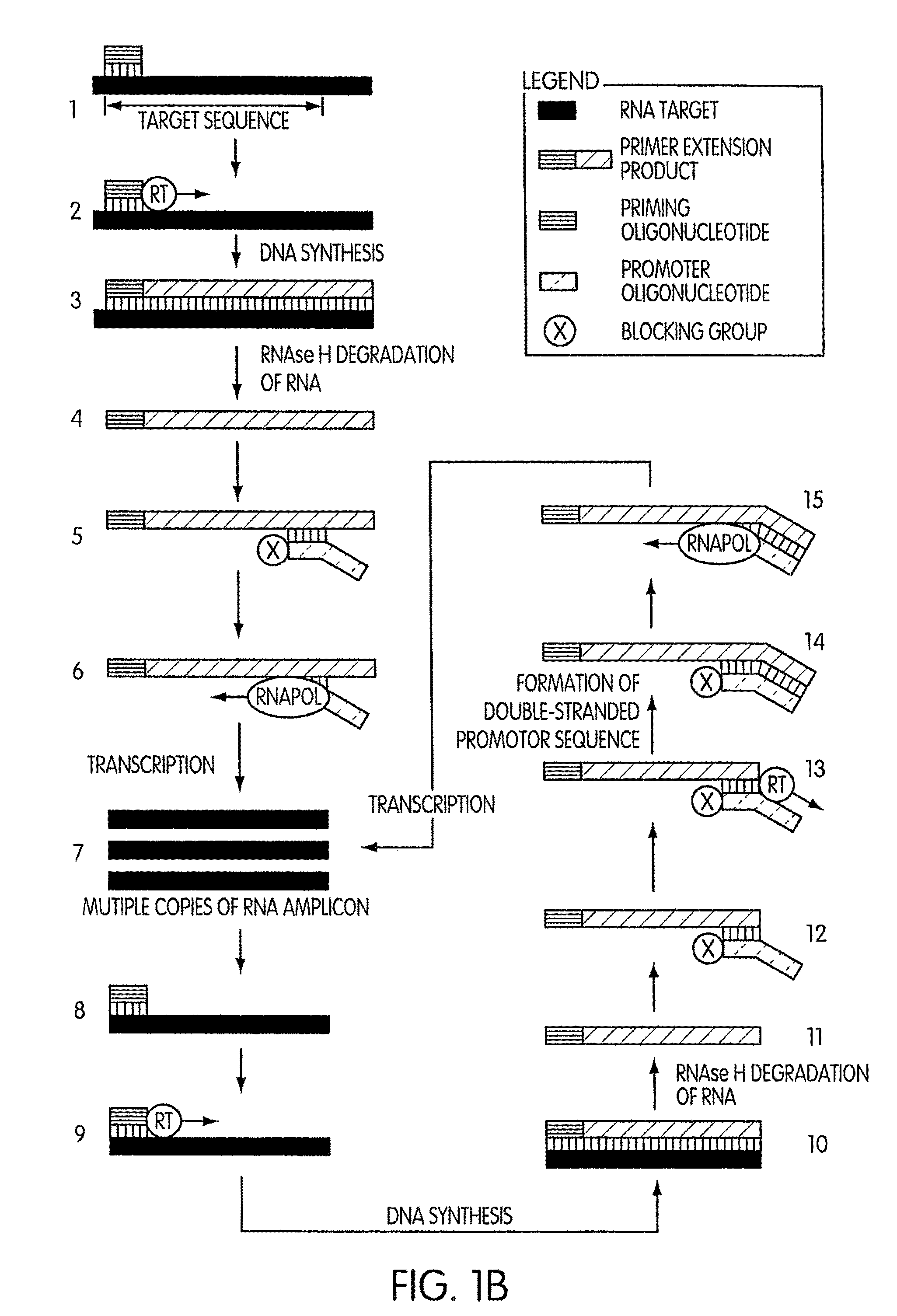
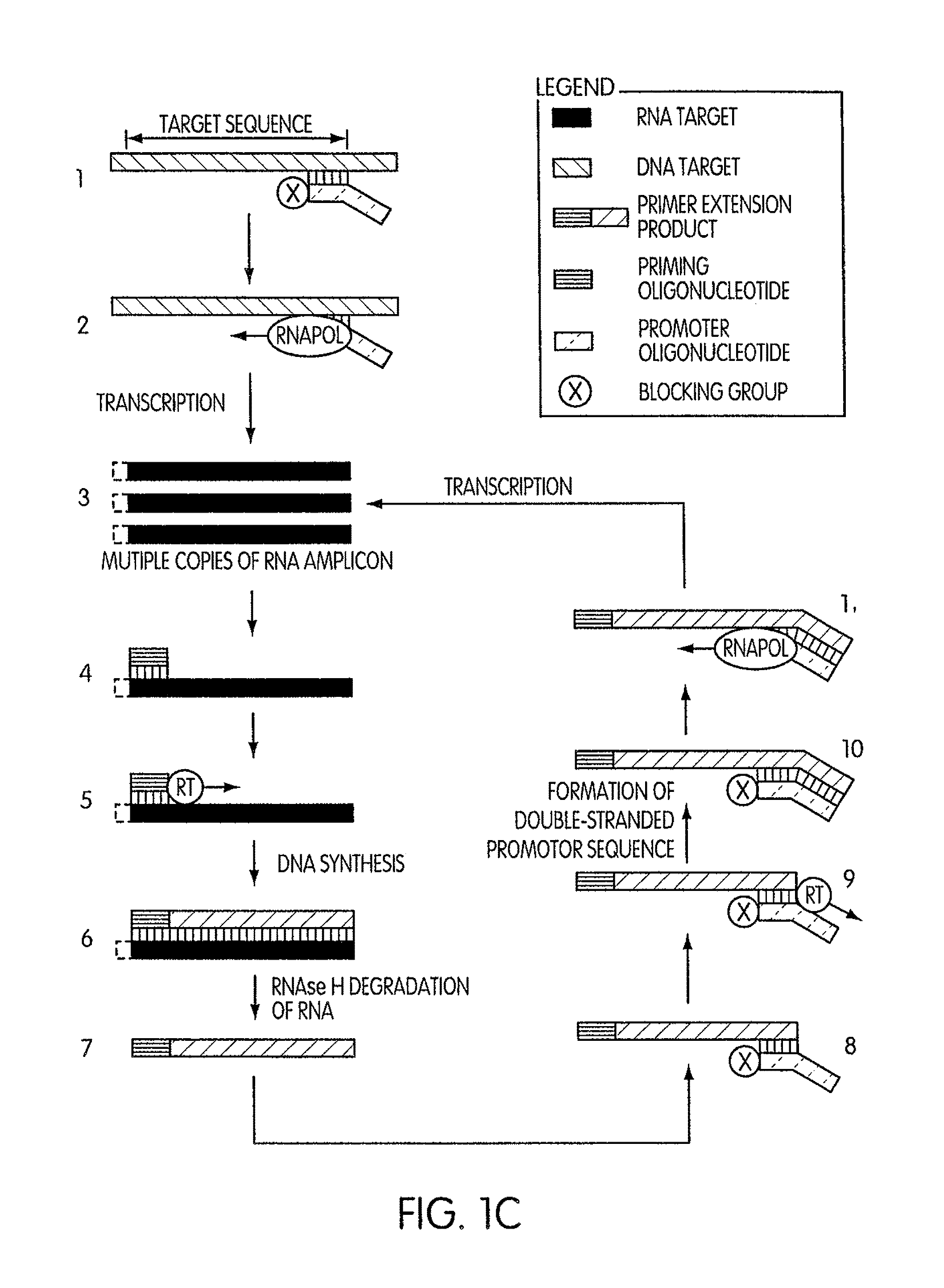
Kits for amplifying DNA which include a priming oligonucleotide that hybridizes to a 3′-end of a DNA target sequence, a displacer oligonucleotide that hybridizes to a target nucleic acid containing the DNA target sequence at a position upstream from the priming oligonucleotide, and a promoter oligonucleotide that includes a region that hybridizes to a 3′-region of a DNA primer extension product that includes the priming oligonucleotide and a promoter for an RNA polymerase. The priming oligonucleotide does not include an RNA region that hybridizes to the target nucleic acid and is selectively degraded by an enzyme activity when hybridized to the target nucleic acid. The kits do not include a restriction endonuclease and oligonucleotides that include a promoter for an RNA polymerase are all modified to prevent the initiation of DNA synthesis therefrom.
Owner:GEN PROBE INC
Method for in vitro transcription using an immobilized restriction enzyme
InactiveUS20180208957A1Enhancing and improving linearizationLow costHydrolasesOn/in organic carrierDNA EndonucleaseRestriction enzyme
The present invention relates to a method for in vitro transcription of a linear template DNA which is produced using an immobilized restriction endonuclease. The invention also relates to mutated restriction enzymes which are suitable for immobilization and a solid support to which these restriction enzymes are immobilized. Further, the present invention relates to an enzyme reactor containing said immobilized restriction endonuclease which enzyme reactor can be used for preparing linearized template DNA. Finally, the present invention relates to the use of said enzyme reactor for preparing a linear template DNA for in vitro transcription. In addition, the present invention relates to a kit comprising the immobilized restriction endonuclease.
Owner:CUREVAC SE
Development of a Marker Foot and Mouth Disease Virus Vaccine Candidate That is Attenuated in the Natural Host
ActiveUS20120315295A1Easy to replaceQuick changeSsRNA viruses positive-senseSugar derivativesSerotypeViral Vaccine
We have generated novel molecularly marked FMDV A24LL3DYR and A24LL3BPVKV3DYR vaccine candidates. The mutant viruses contain a deletion of the leader coding region (LL) rendering the virus attenuated in vivo and negative antigenic markers introduced in one or both of the viral non-structural 3Dpol and 3B proteins. The vaccine platform includes unique restriction endonuclease sites for easy swapping of capsid proteins for different FMDV subtypes and serotypes. The mutant viruses produced no signs of FMD and no shedding of virulent virus in cattle. No clinical signs of disease or fever were observed and no transmission to in-contact animals was detected in pigs inoculated with live A24LL3DYR. Cattle immunized with chemically inactivated vaccine candidates showed an efficacy comparable to a polyvalent commercial FMDV vaccine. These vaccine candidates used in conjunction with a cELISA provide a suitable target for DIVA companion tests.
Owner:US SEC AGRI
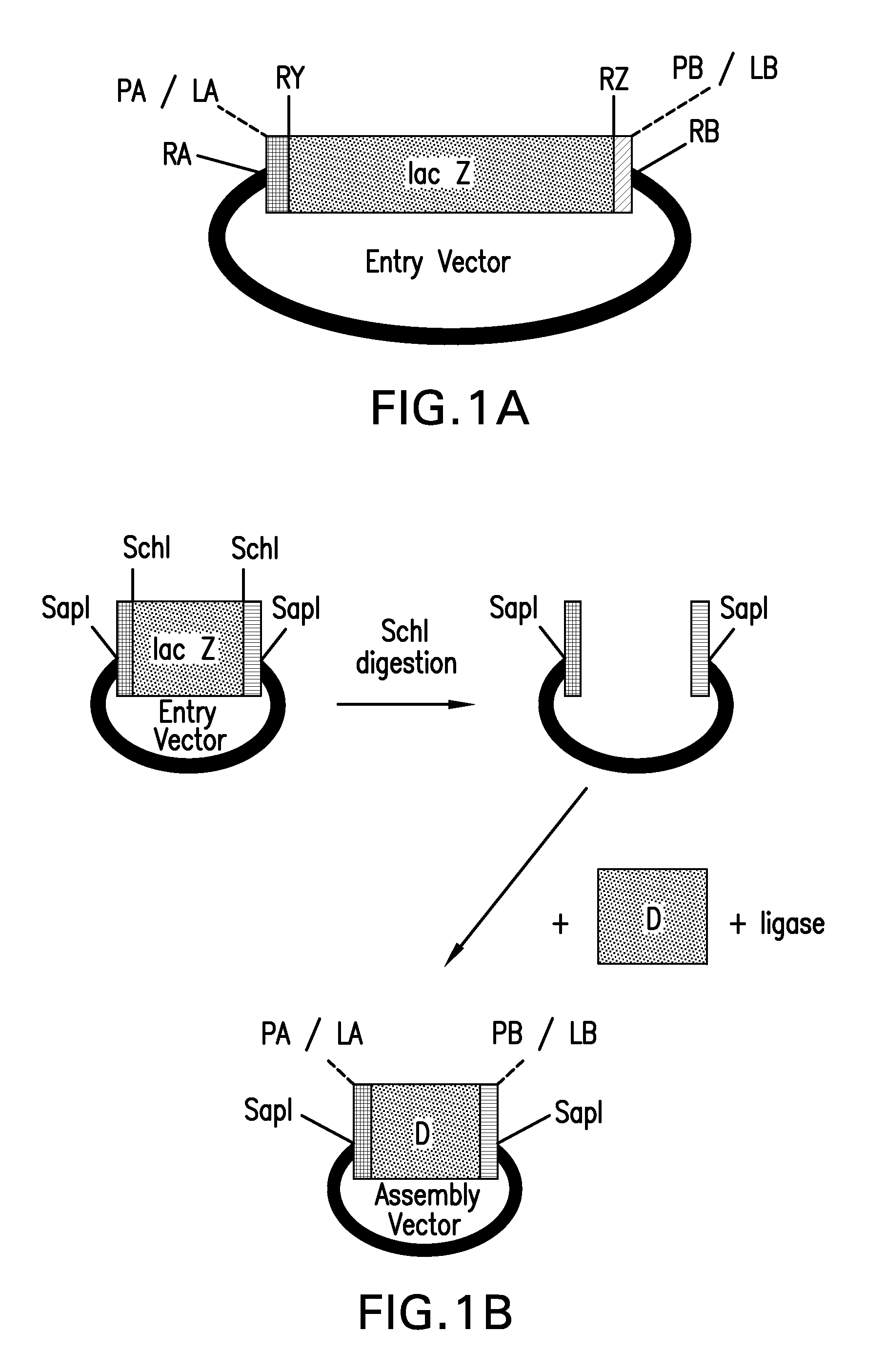
Compositions and methods for the assembly of polynucleotides
ActiveUS20100124768A1Efficient assemblyHigh melting temperatureFungiSugar derivativesNucleotidePolymerase L
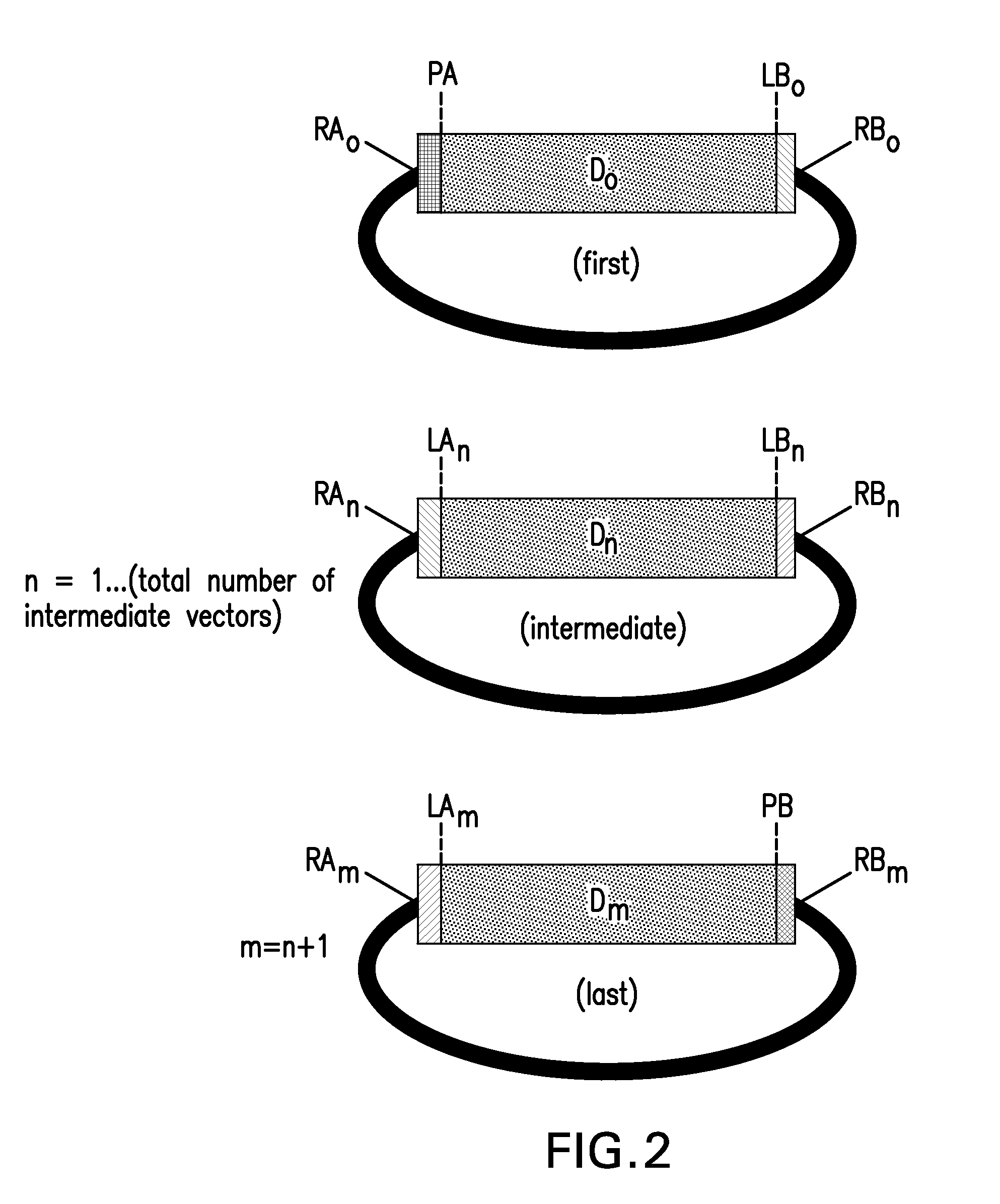
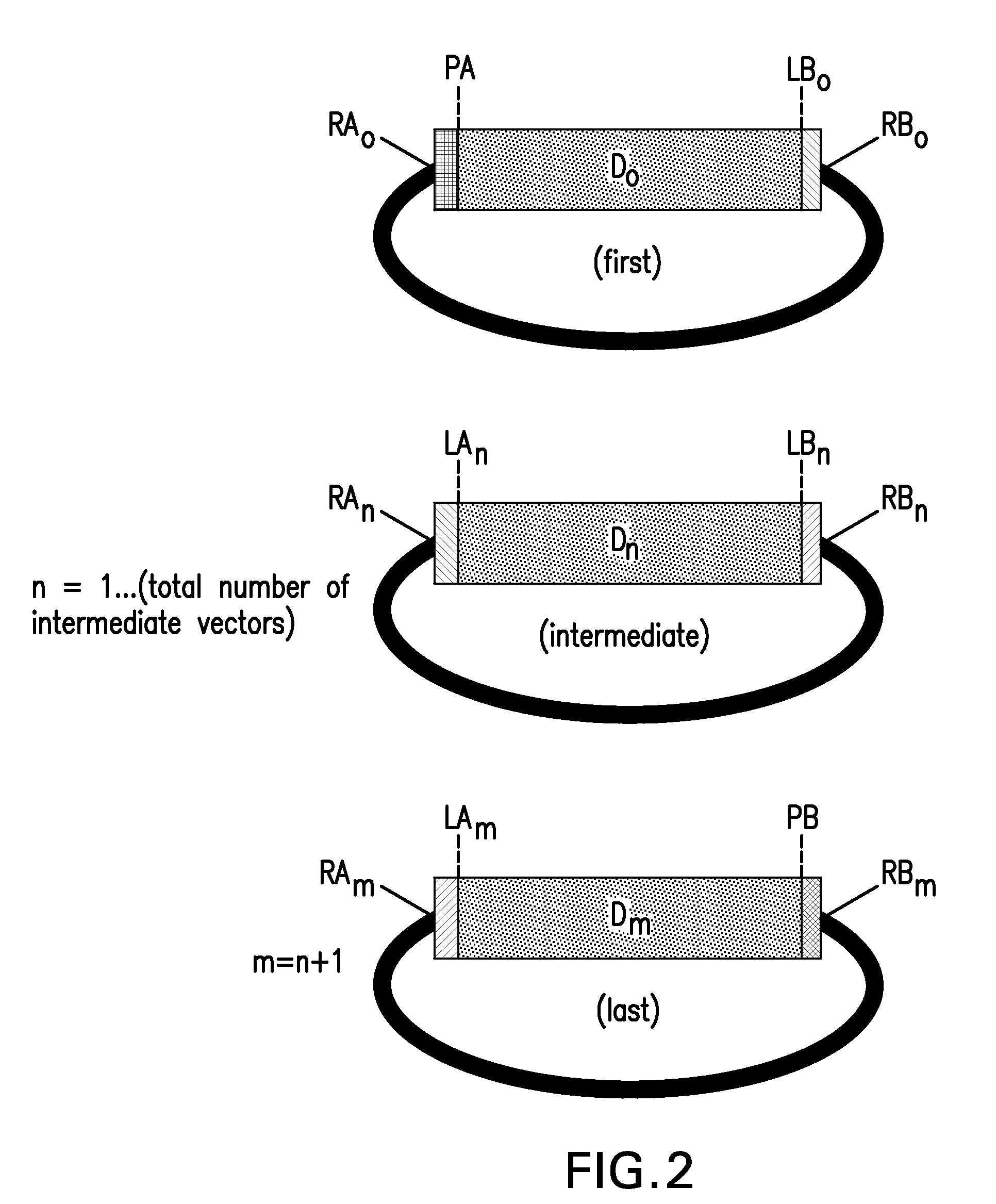
The present invention provides compositions and methods for rapid assembly of one or more assembled polynucleotides from a plurality of component polynucleotides. The methods of the invention utilize circular nucleic acid vectors that comprise a DNA segment D flanked by an annealable linker sequence, annealable linker sequence pairs LA and LB, or annealable linker sequence / primer binding segment pairs LA and PB or PA and LB. Restriction endonuclease digestion of a plurality of vectors containing the DNA segments to be assembled generates a plurality of DNA fragments comprising the elements PA-D-LB, LA-D-LB, and LA-D-PB or D-LB, LA-D-LB, and LA-D. The sequences of annealable linker sequences LA and LB provide complementary termini to the DNA fragments, which are utilized in host cell mediated homologous recombination or together with primer binding segments PA and PB in a polymerase cycling assembly reaction for the ordered assembly of the various DNA segments into one or more assembled polynucleotides.
Owner:AMYRIS INC
Compositions and methods for the assembly of polynucleotides
ActiveUS20100136633A1Efficient assemblyHigh melting temperatureFungiSugar derivativesNucleotidePolymerase L
The present invention provides compositions and methods for rapid assembly of one or more assembled polynucleotides from a plurality of component polynucleotides. The methods of the invention utilize circular nucleic acid vectors that comprise a DNA segment D flanked by an annealable linker sequence, annealable linker sequence pairs LA and LB, or annealable linker sequence / primer binding segment pairs LA and PB or PA and LB. Restriction endonuclease digestion of a plurality of vectors containing the DNA segments to be assembled generates a plurality of DNA fragments comprising the elements PA-D-LB, LA-D-LB, and LA-D-PB or D-LB, LA-D-LB, and LA-D. The sequences of annealable linker sequences LA and LB provide complementary termini to the DNA fragments, which are utilized in host cell mediated homologous recombination or together with promer binding segments PA and PB in a polymerase cycling assembly reaction for the ordered assembly of the various DNA segments into one or more assembled polynucleotides.
Owner:AMYRIS INC
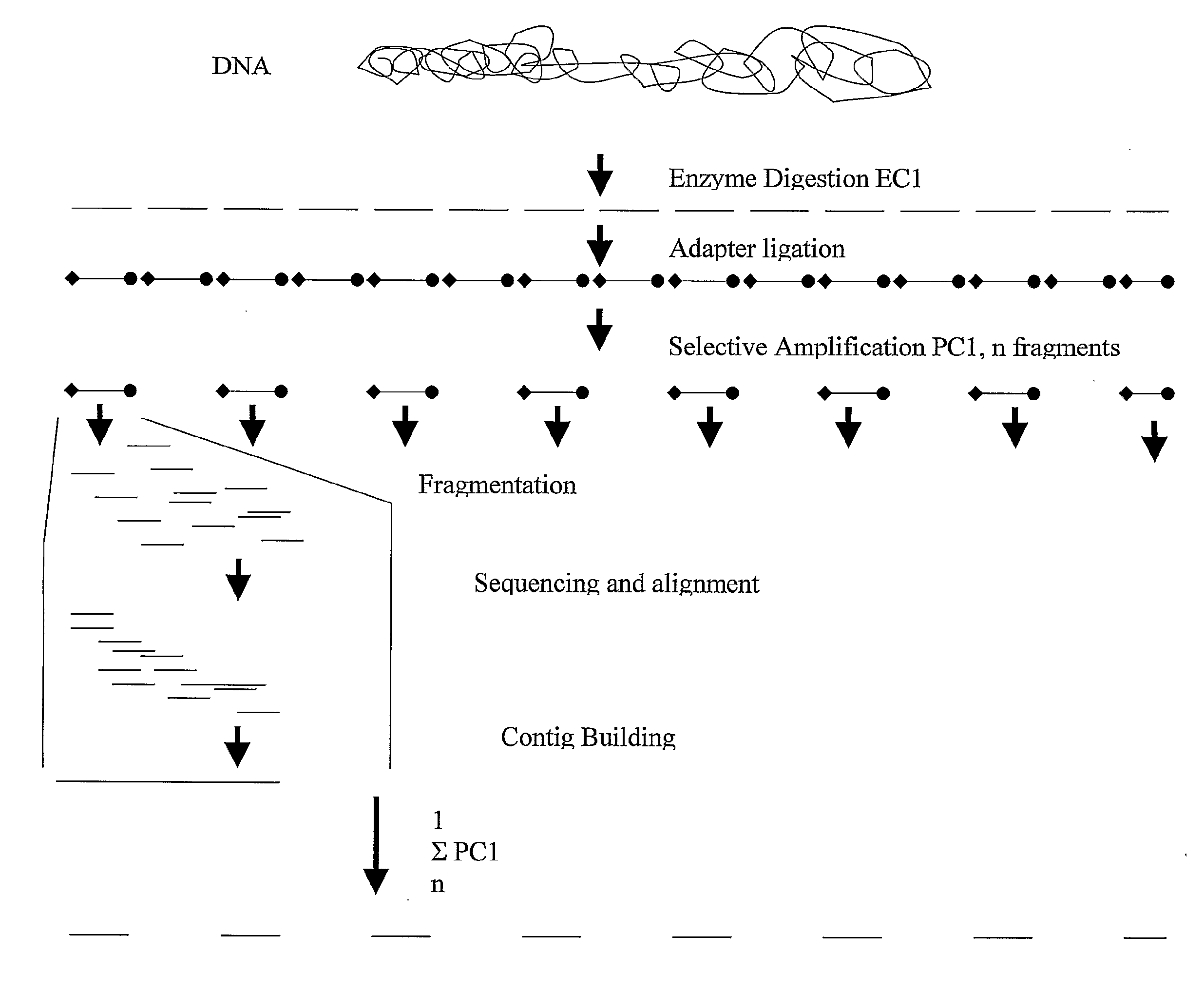
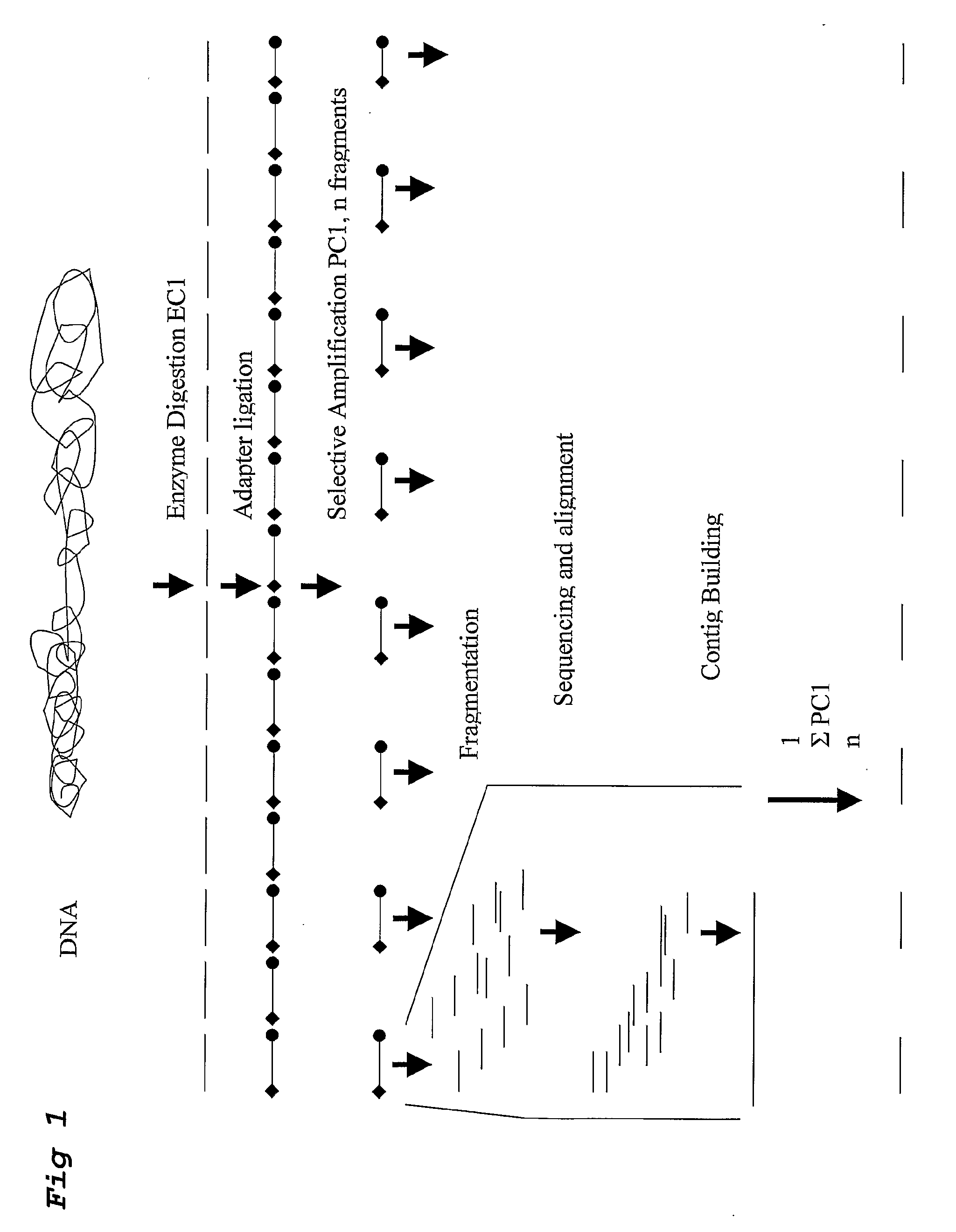
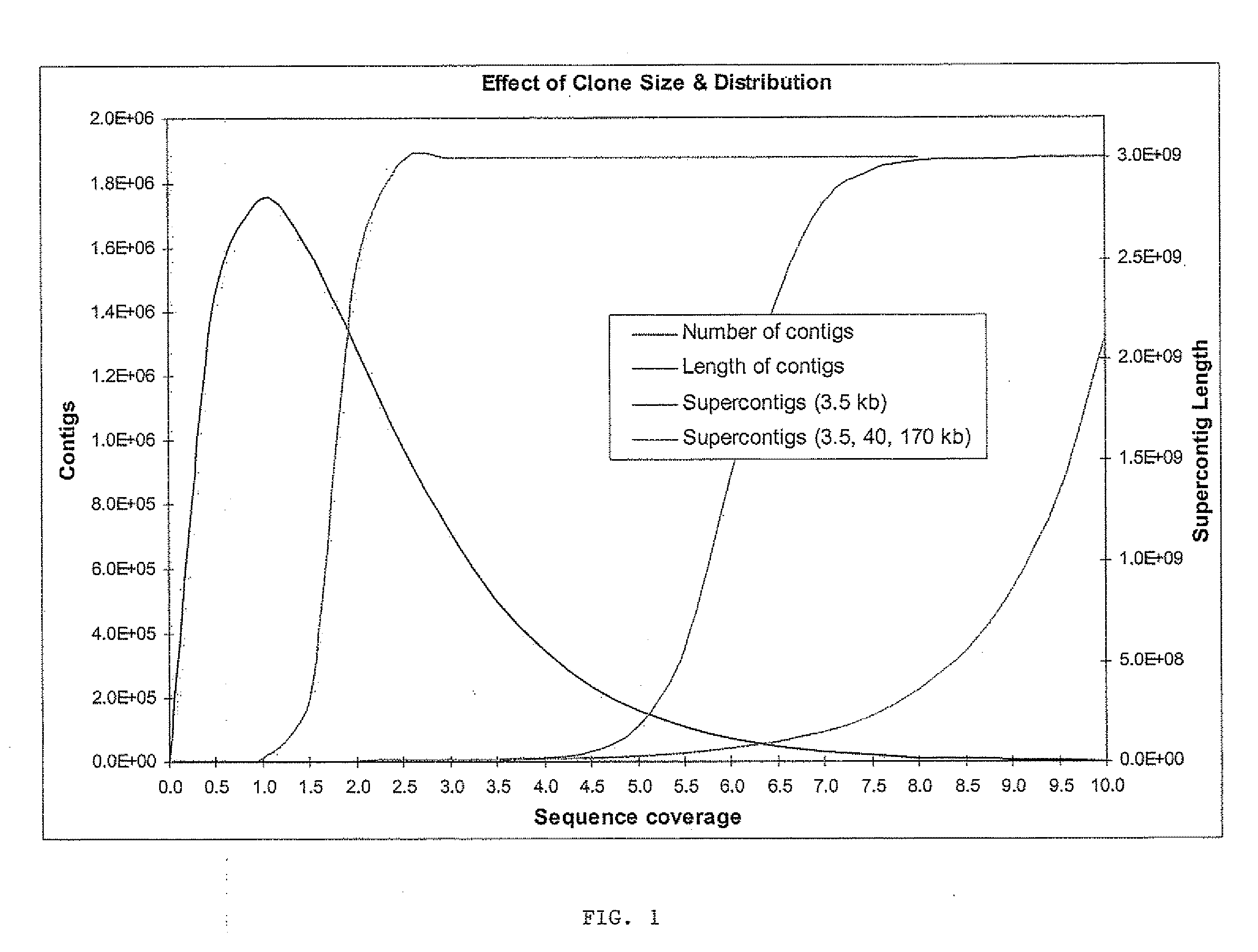
Strategies for sequencing complex genomes using high throughput sequencing technologies
InactiveUS20090142758A1Easy to useQuick sortingMicrobiological testing/measurementContigNucleotide sequencing
A method for determining a genome sequence comprising the steps of digesting the genome with at least one first restriction endonuclease, ligating at least one adaptor to the restriction fragments of the first subset, selectively amplifying the first set of adaptor-ligated restriction fragments using a first primer combination wherein at least a first primer contains a first selected sequence at the 3′ end of the primer sequence, comprising 1-10 selective nucleotides, repeating these steps with at least a second primer combinations wherein the primer contains a different second selected sequence, fragmenting each of the subsets of amplified adaptor-ligated restriction fragments to generate sequencing libraries, determine the nucleotide sequence of the fragments, aligning the sequence of the fragments in each of the libraries to generate contigs, repeating these steps for one second and / or further restriction endonucleases, aligning the contigs obtained for each of the second and / or further restriction endonucleases to provide for a sequence of the genome.
Owner:KEYGENE NV
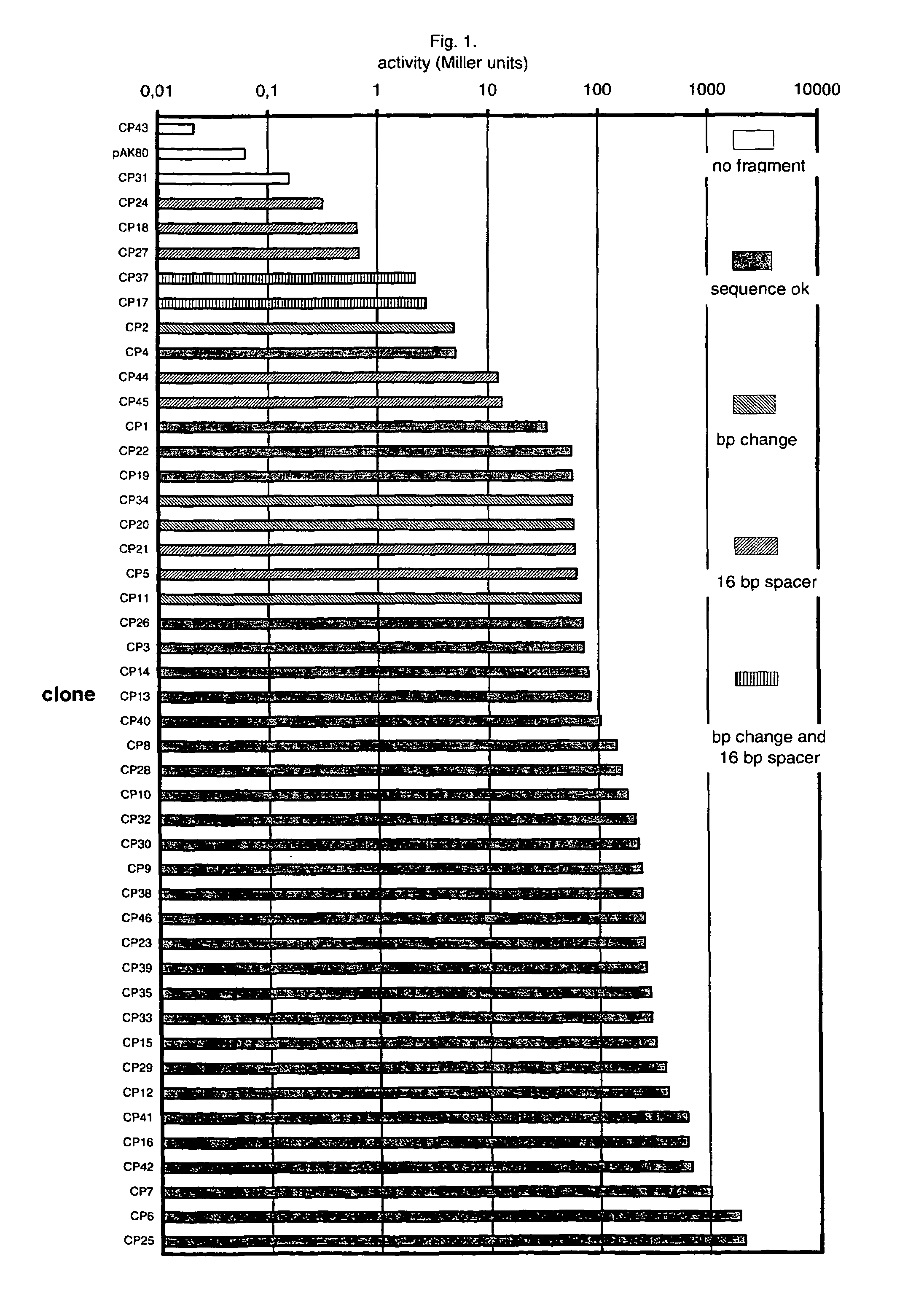
Artificial promoter libraries for selected organisms and promoters derived from such libraries
InactiveUS7199233B1Strong impactNone is suitable for purposeSugar derivativesMicrobiological testing/measurementNucleotideNucleobase
An artificial promoter library (or a set of promoter sequences) for a selected organism or group of organisms is constructed as a mixture of double stranded DNA fragments, the sense strands of which comprise at least two consensus sequences of efficient promoters from said organism or group of organisms, or parts thereof comprising at least half of each, and surrounding intermediate nucleotide sequences (spacers) of variable length in which at least 7 nucleotides are selected randomly among the nucleobases A, T, C and G. The sense strands of the double stranded DNA fragments may also include a regulatory DNA sequence imparting a specific regulatory feature, such as activation by a change in the growth conditions, to the promoters of the library. Further, they may have a sequence comprising one or more recognition sites for restriction endonucleases added to one or both of their ends. The selected organism or group of organisms may be selected from prokaryotes and from eukaryotes; and in prokaryotes the consensus sequences to be retained most often will comprise the −35 signal (−35 to −30): TTGACA and the −10 signal (−12 to −7): TATAAT or parts of both comprising at least 3 conserved nucleotides of each, while in eukaryotes said consensus sequences should comprise a TATA box and at least one upstream activation sequence (UAS). Such artificial promoter libraries can be used, e.g., for optimizing the expression of specific genes in various selected organisms.
Owner:JENSEN PETER RUHDAL +1
Fluorometric assay for detecting nucleic acid cleavage
A method of detecting an enzyme-mediated DNA cleavage reaction in a fluorometric assay is provided. The method can be used to detect DNA cleavage caused by restriction endonucleases, retroviral integrase enzymes, DNases, RNases, or enzymes utilized in other strand separating processes in molecular biology.
Owner:GEORGETOWN UNIV
Method for engineering strand-specific nicking endonucleases from restriction endonucleases
ActiveUS7943303B2Efficiently obtainedImprove efficiencySugar derivativesHydrolasesSingle strandIn vivo
Owner:NEW ENGLAND BIOLABS
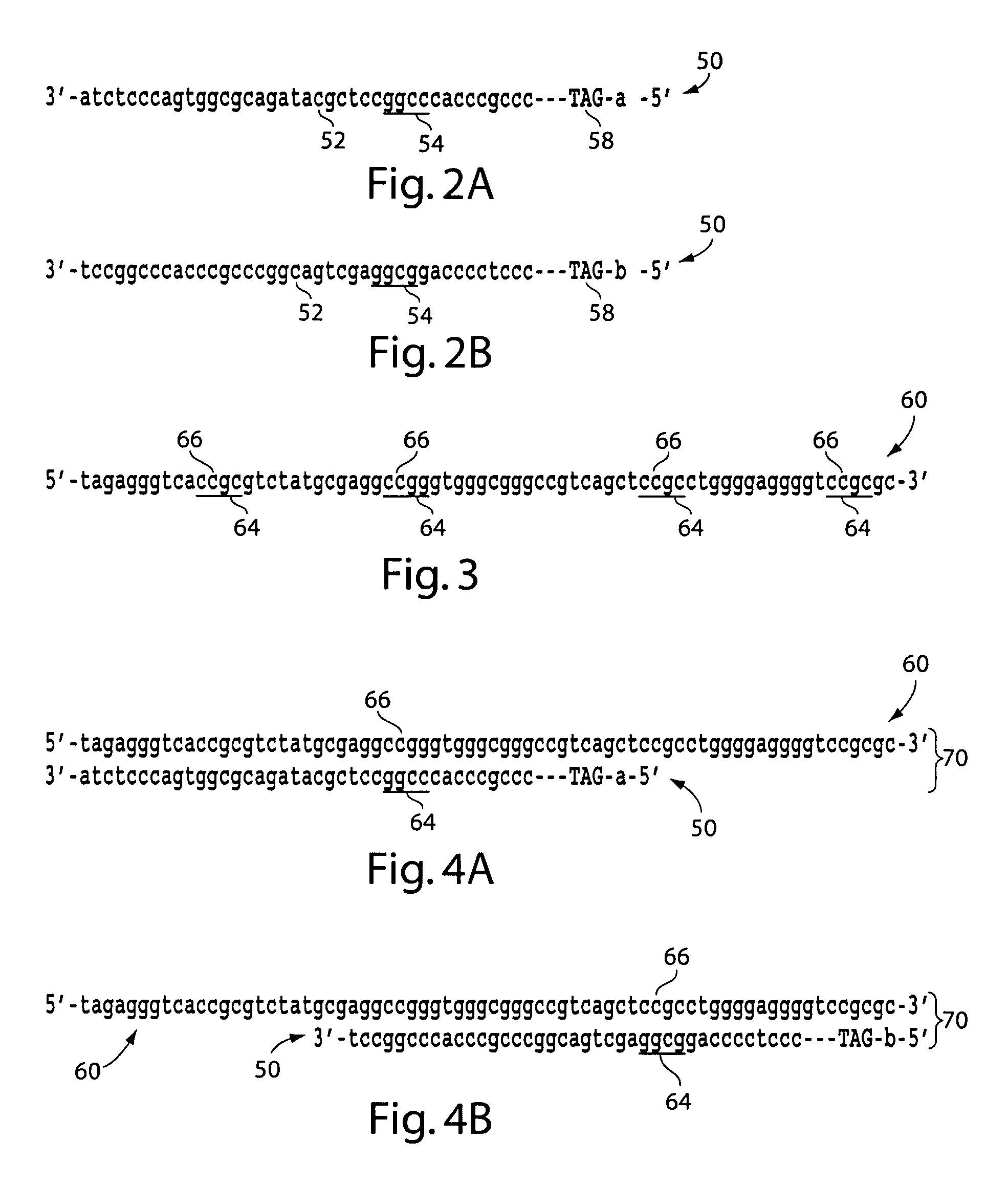
Methods for producing a paired tag from a nucleic acid sequence and methods of use thereof
Methods for producing a paired tag from a nucleic acid sequence are provided in which the paired tag comprises the 5′ end tag and 3′ end tag of the nucleic acid sequence. In one embodiment, the nucleic acid sequence comprises two restriction endonuclease recognition sites specific for a restriction endonuclease that cleaves the nucleic acid sequence distally to the restriction endonuclease recognition sites. In another embodiment, the nucleic acid sequence further comprises restriction endonuclease recognition sites specific for a rare cutting restriction endonuclease. Methods of using paired tags are also provided. In one embodiment, paired tags are used to characterize a nucleic acid sequence. In a particular embodiment, the nucleic acid sequence is a genome. In one embodiment, the characterization of a nucleic acid sequence is karyotyping. Alternatively, in another embodiment, the characterization of a nucleic acid sequence is mapping of the sequence. In a further embodiment, a method is provided for identifying nucleic acid sequences that encode at least two interacting proteins.
Owner:APPL BIOSYSTEMS INC
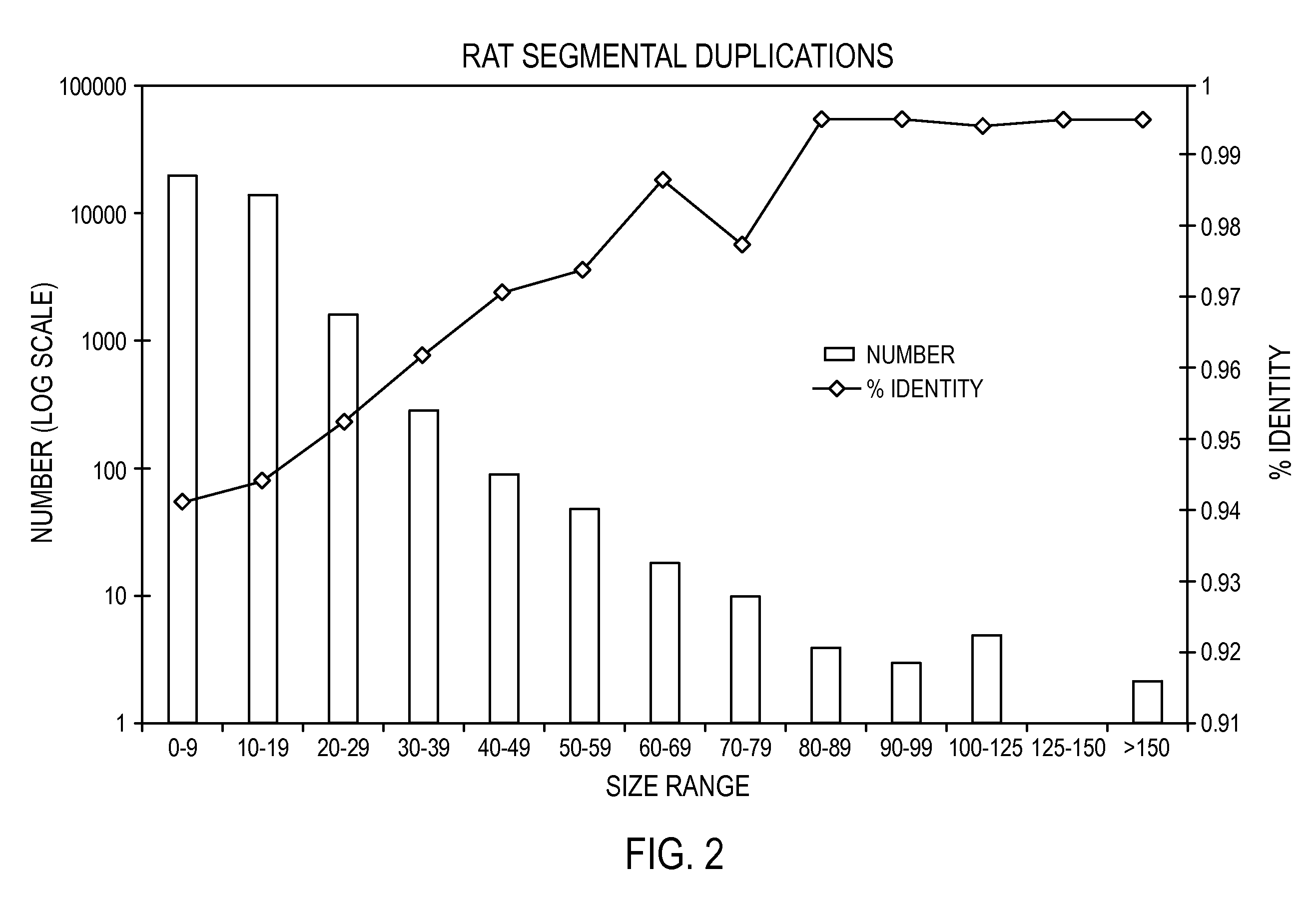
Methods for Nucleic Acid Mapping and Identification of Fine Structural Variations in Nucleic Acids
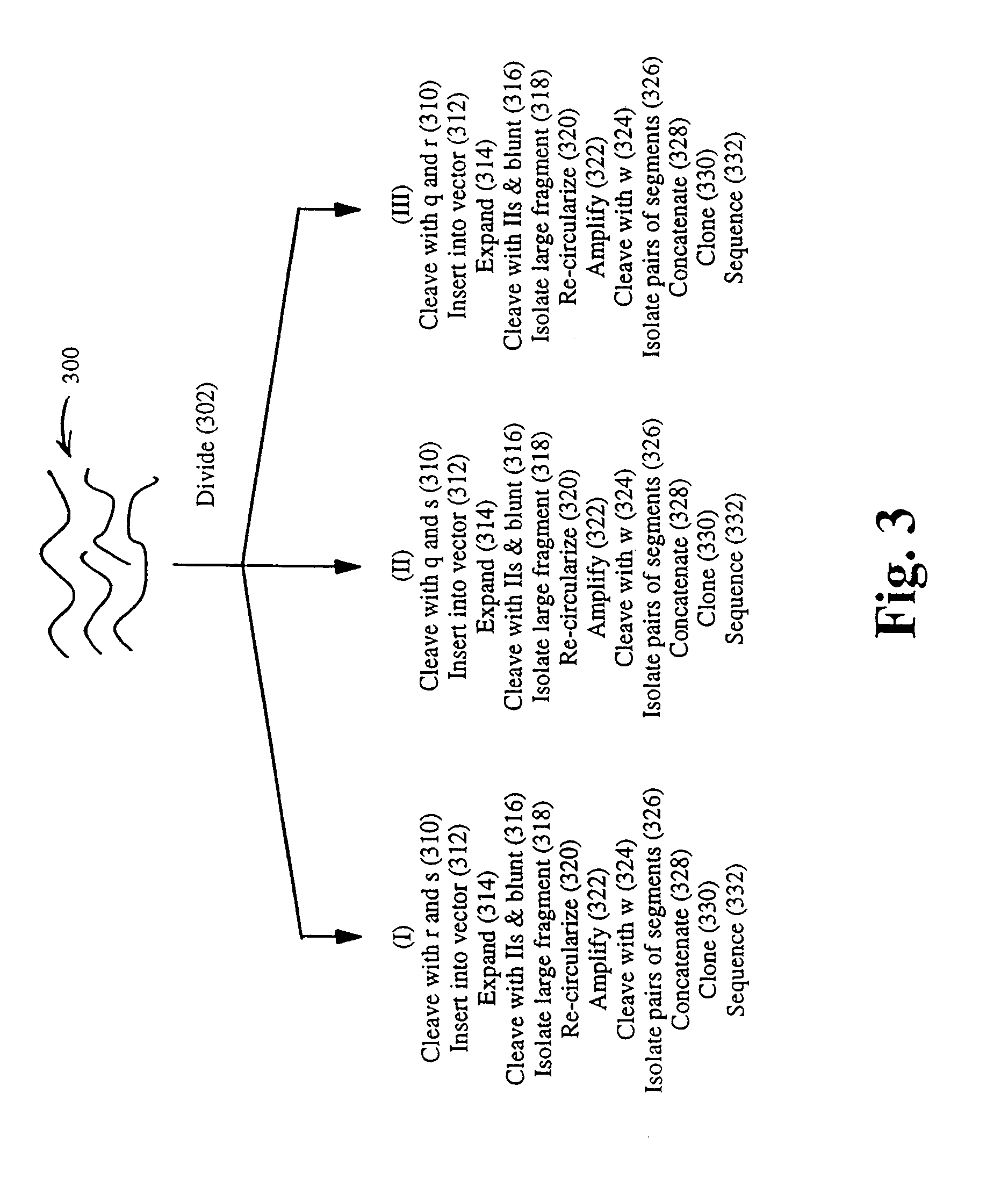
InactiveUS20090156431A1Increase insert size carrying capacityQuickly reconfiguredVector-based foreign material introductionLibrary creationSmaI restriction endonucleaseNucleic acid molecule
An in vitro, extracellular method of juxtaposing sequence tags (GVTs) where two constituent members of a tag pair (GVT-pair) are unique positional markers of a defined separation distance and / or are markers of nucleic acid positions that demarcate adjacent cleavage sites for one or more different restriction endonucleases along the length of a plurality of target nucleic acid molecules, the method comprising: Fragmenting the target nucleic acid molecule to form target DNA insert; ligating a DNA adaptor having one or more restriction endonuclease recognition sites to both ends of a fragmented target DNA insert and the ligation of the adaptor-ligated target DNA insert to a DNA backbone to create a circular molecule; digesting the adaptor using a restriction endonuclease at the recognition site to cleave the target DNA insert at a defined distance from each end thereof to create two sequence tags (GVTs) comprising terminal sequences of the target DNA insert that are attached to the linear DNA backbone; and recircularizing the linear DNA backbone with the attached GVTs to obtain a circular DNA molecule including a GVT pair having two juxtaposed GVTs; GVT-pair DNA is recovered by nucleic acid amplification.
Owner:LOK SI
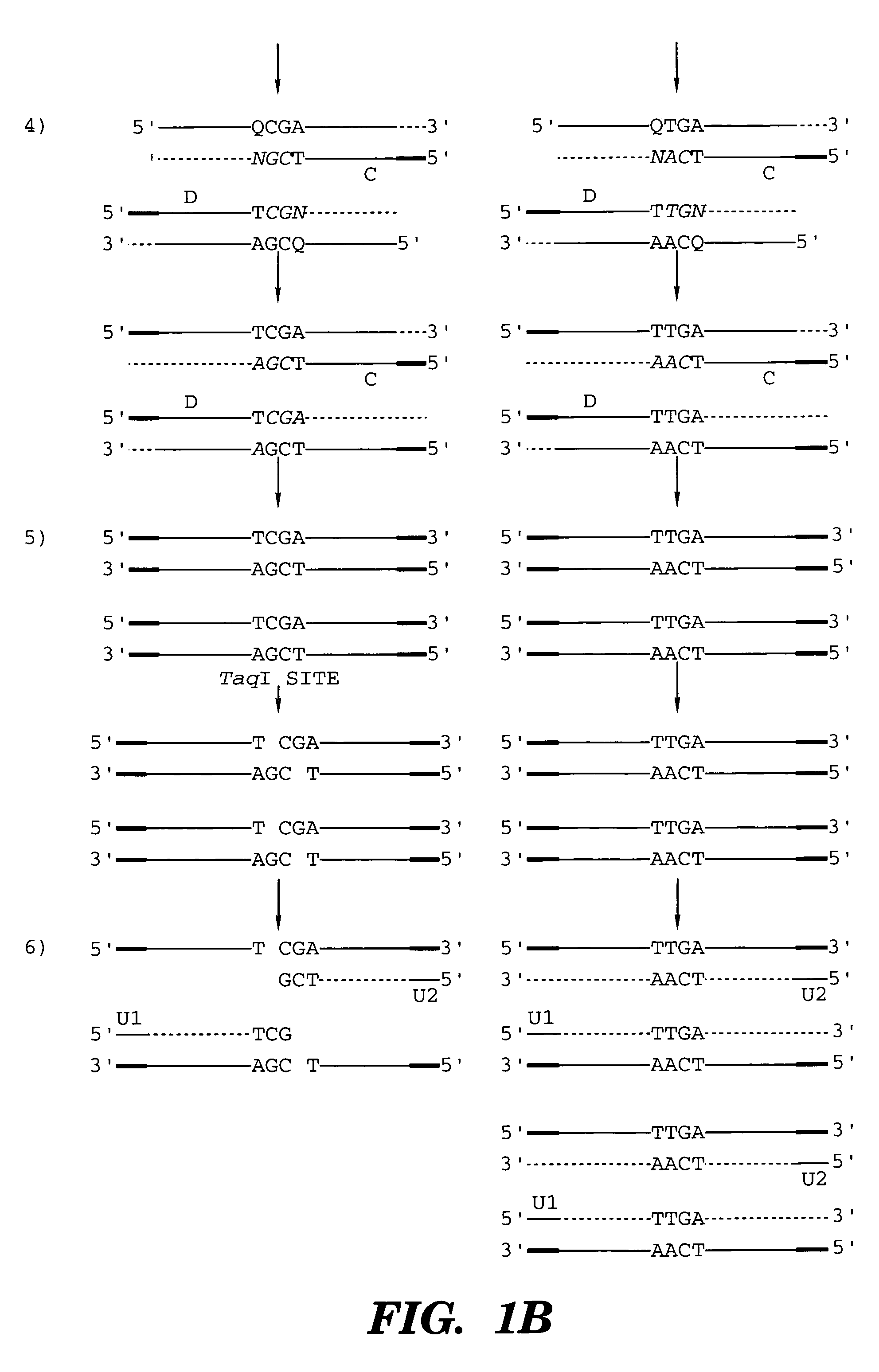
Restriction endonucleases and their uses
A restriction endonuclease with a recognition sequence 5′-TCGA-3′. The restriction endonuclease is sensitive to the presence of a modified cytosine residue in the recognition sequence. Methods and kits using the restriction endonuclease with a recongition sequence 5′-TCGA-3′ are also disclosed.
Owner:THERMO FISHER SCI BALTICS UAB
Multiple sequencible and ligatible structures for genomic analysis
InactiveUS7262030B2Highly cost-effectiveImprove throughputMicrobiological testing/measurementFermentationNon targetedGenome
High throughput methods and kits for single nucleotide polymorphism (SNP) genotyping are provided. The methods involve utilizing nested PCR amplification reactions which produce sequencible and ligatible structures. An outer PCR primer set amplifies the SNP, and an inner PCR primer set amplifies a portion of the DNA amplified by the outer primer set, but does not amplify the SNP itself. The inner and outer primers may reaction include non-target common domain sequences, and the inner primer common domain sequences may comprise digestion restriction endonuclease recognition sites. The design of the inner primer set allows precise tailoring of the sequencible and ligatible structures with respect to length and base composition.
Owner:VIRGINIA COMMONWEALTH UNIV
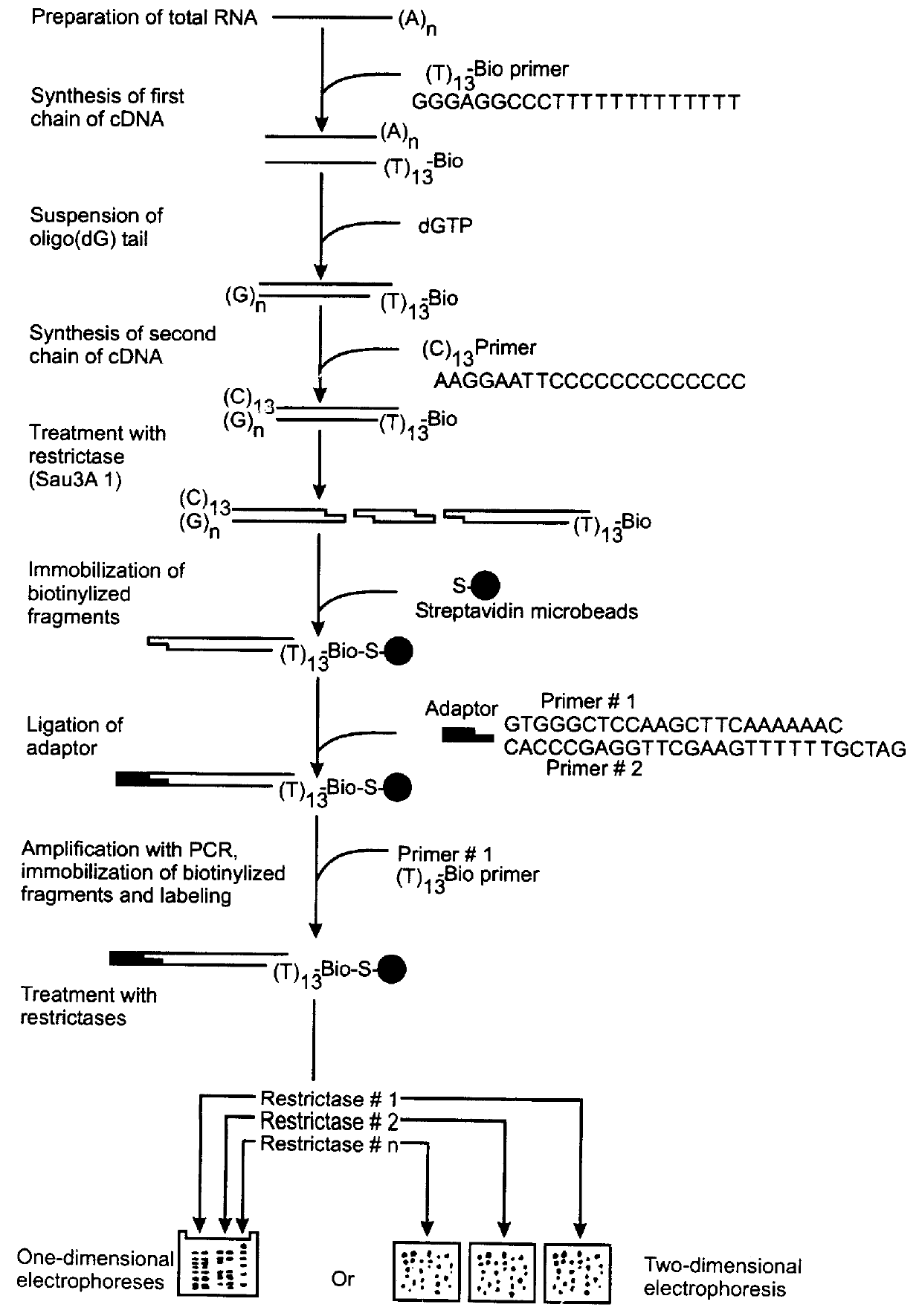
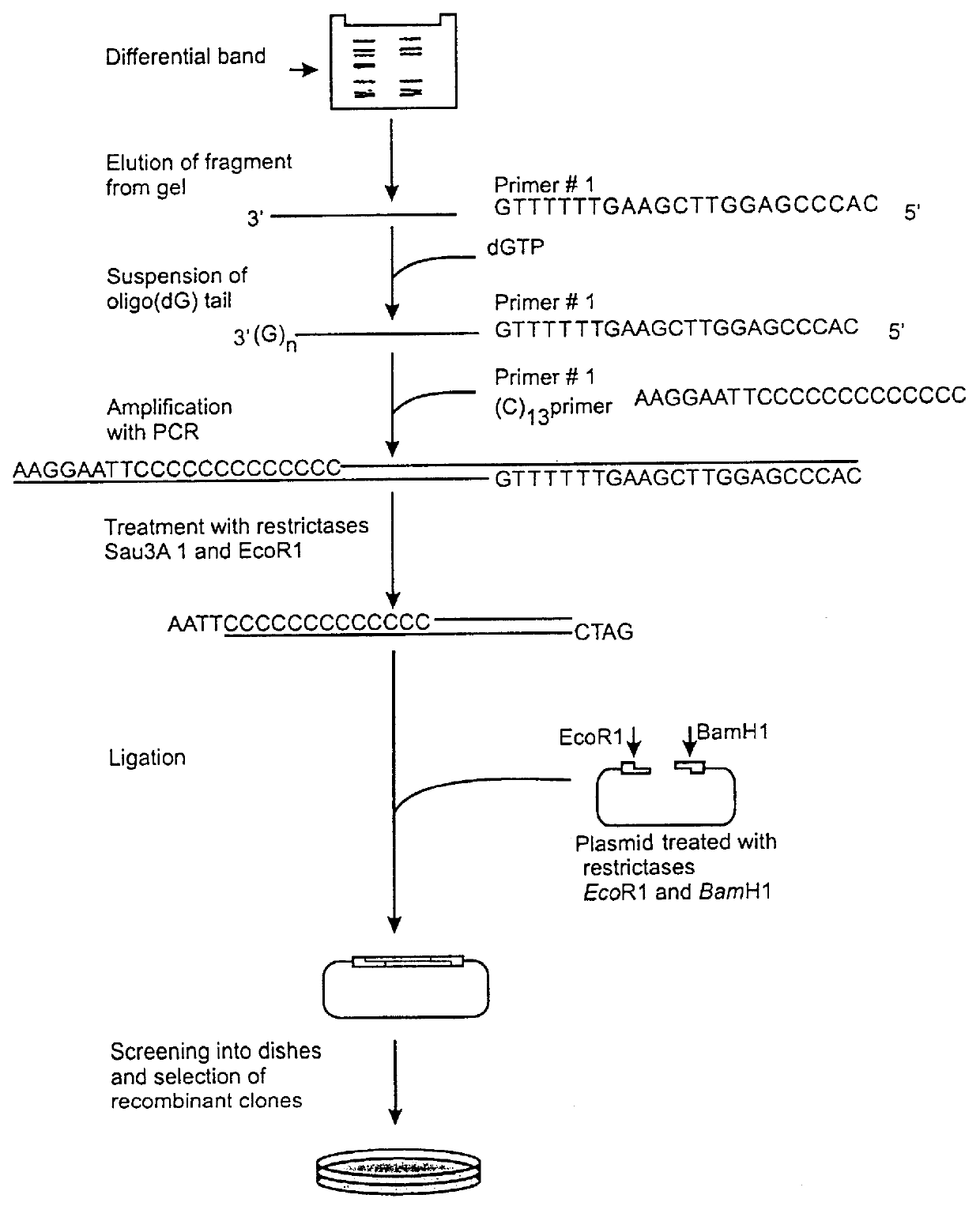
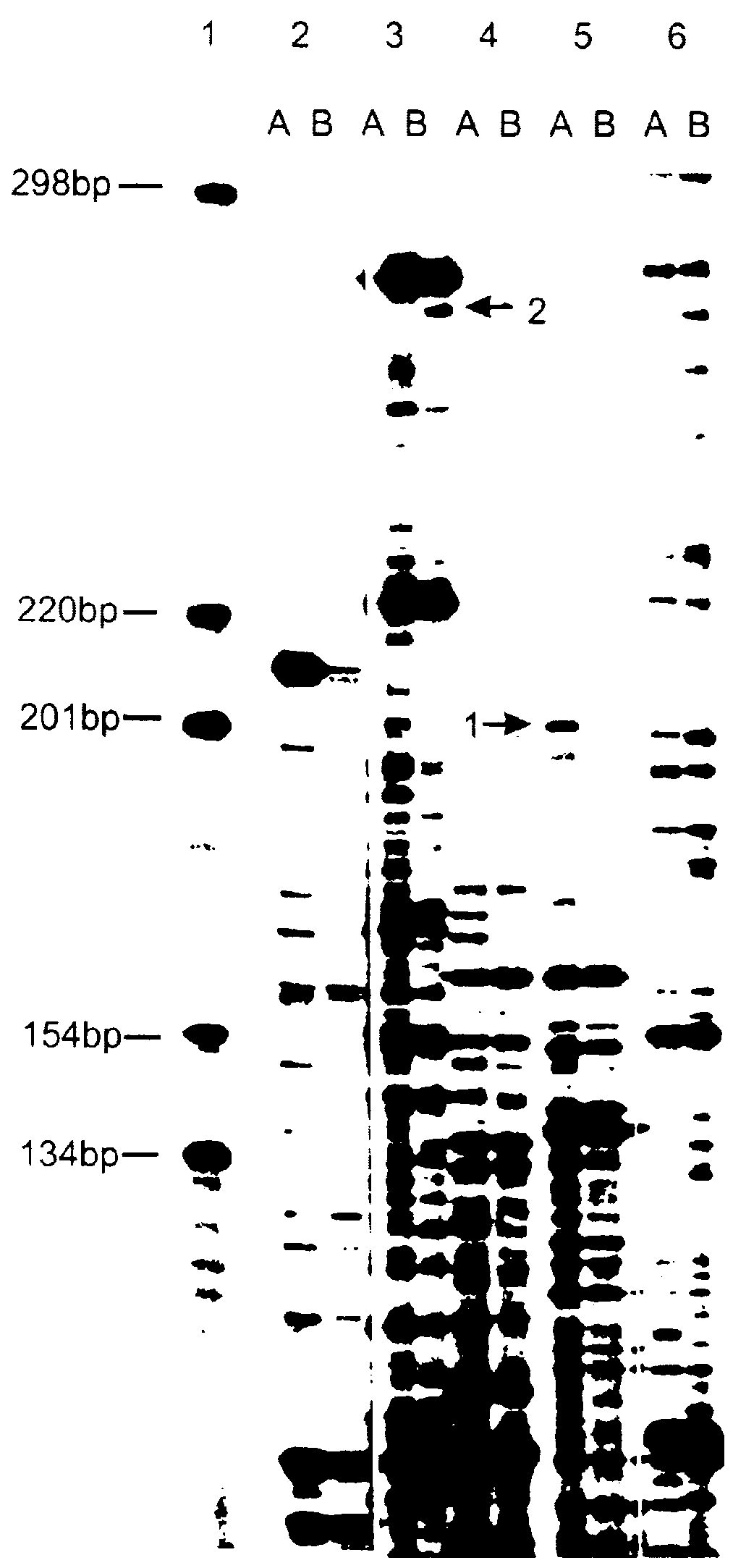
Method of identification and cloning differentially expressed messenger RNAs
A method of identification of differentially expressed messenger RNA (mRNA) which consists of synthesizing from a set of sequences of mRNA sets of fragments of complementary DNA (cDNA), which are separated with the aid of gel electrophoresis and the pictures of separation of the cDNA from different types of cells are compared and fragments with differential signal intensity are identified. For formation of the set of fragments the cDNA is cleaved with the aid of restriction nucleases. A method of cloning of differentially expressed mRNAs consists of synthesizing from sets of sequences of mRNAs from different types of cells sets of fragments of complementary DNA (cDNA) which are separated with the aid of gel electrophoresis, the pictures of the separation of the cDNA from different types of cells are compared, fragments of cDNA with different signal intensities are separated from the gel, amplified with the aid of a polymerase chain reaction and cloned to a plasmid or phage vector. For the formation of the set of fragments one carries out cleavage of the cDNA with the aid of restriction endonucleases and uses only those fragments of cDNA that correspond to the 3' or 5' end regions of the mRNAs.
Owner:NEW YORK BLOOD CENT
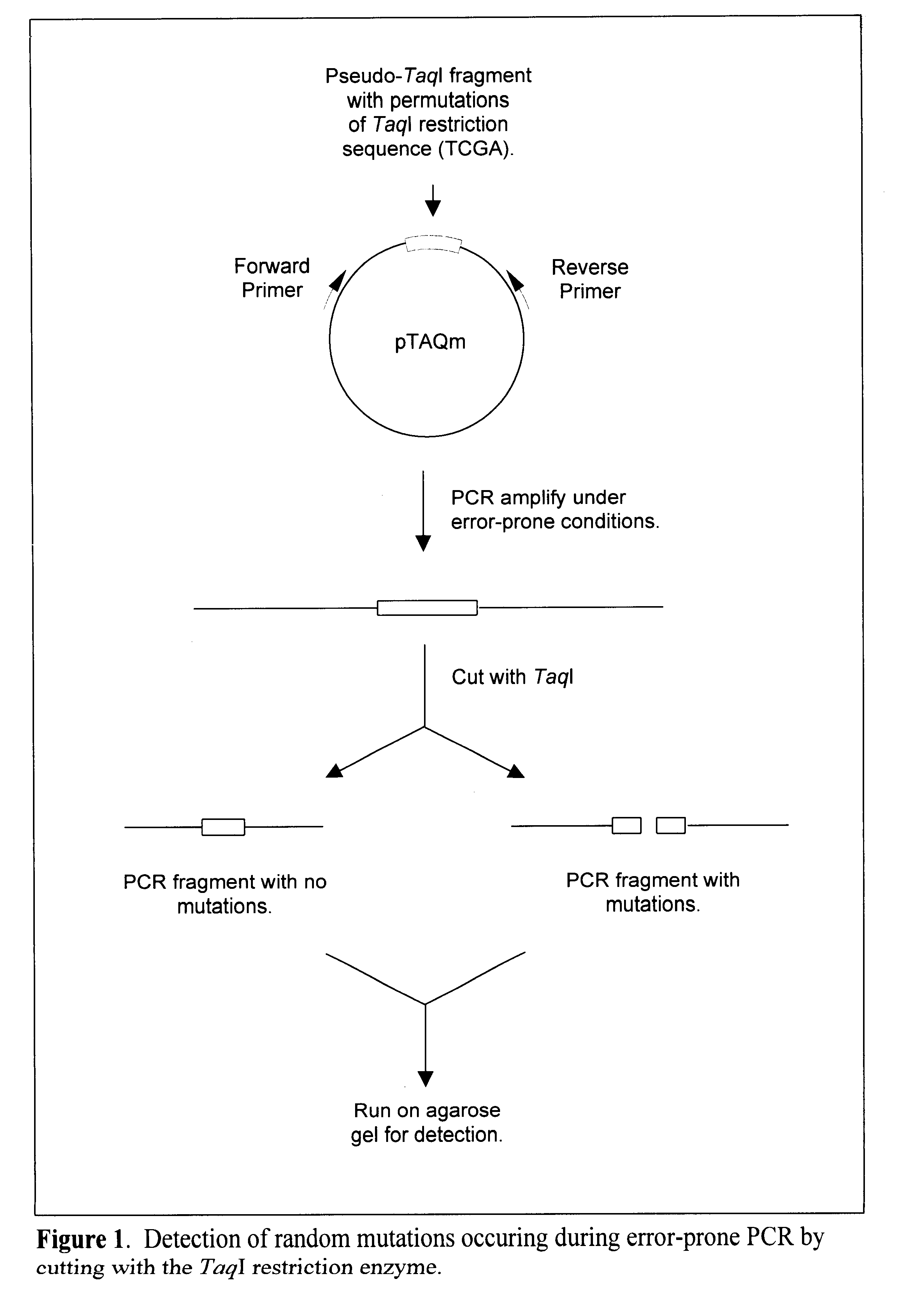
Methods and kits for determining the fidelity of polymerase chain reaction conditions
Methods are provided for evaluating the fidelity of a given set of polymerase chain reaction conditions. In the subject methods, a template polydeoxyribonucleotide is amplified under the to be evaluated polymerase chain reaction conditions, where the template polydeoxyribonucleotide includes a pseudo restriction endonuclease restriction site. The resultant amplified product population is then contacted with the corresponding restriction endonuclease and resultant cleavage products, if any, are detected. The fidelity of the polymerase chain reaction conditions is then derived from the detected cleavage products (or absence thereof). Also provided are kits for use in practicing the subject methods. The subject methods are suited for determining the fidelity of a given polymerase under PCR conditions, and are particularly suited for determining the fidelity of a thermostable polymerase under PCR conditions.
Owner:CLONTECH LAB
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com