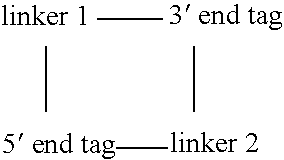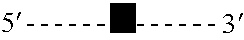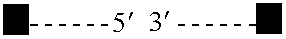Methods for producing a paired tag from a nucleic acid sequence and methods of use thereof
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0098] The whole genome shotgun sequencing, assembly and finishing paradigm is generally the strategy of choice for microbial and fungal genome sequencing. Contrary to the expectations of some, recent advances in the development and application of whole genome assembly (WGA) software, such as Arachne, the Celera Assembler, Phusion, or Jazz, have also demonstrated that it is straightforward to produce a high quality assembly of plant and mammalian genomes using such tools (Mural, Adams et al. 2002; Jaffe, Butler et al. 2003). The cost-advantages and simplicity of the whole genome approach relative to a BAC based or hybrid sequencing strategy argue strongly for its continued development and application in future sequencing projects. One problem with the BAC based approaches is the high cost and operational burden associated with the production of 15,000-25,000 individual BAC subclone libraries, the 15-20% waste associated with re-sequencing the vector as well as ...
example 2
Paired Genome Sequence Tag Method
[0118] Typically, a collection of genomic DNA fragments is produced by fragmentation using one or more restriction endonucleases, or by shearing, with subsequent repair of the ends to produce blunt-ended fragments. Fragments of a specific size range are purified by gel electrophoresis, followed by extraction from a gel slice by electroelution, extraction with chaotropic salts and glass beads, or enzymatic dissolution of the gel followed by phenol extraction and ethanol precipitation. The fragments are ligated to an adapter containing a type IIs restriction enzyme site juxtaposed to a blunt end and the resulting circularized products are subsequently digested with the enzyme to create short tags specific to the ends of the DNA fragment. These tags, which remain connected to each other through the process via the adapter are ligated to form concatemers of paired tags. The concatemers are sequenced and the paired tags extracted from the sequence. The p...
example 3
Paired Tag Two-Hybrid
[0121] This method provides a linked pair of bait and prey molecules that derive from cells that screen positive for an interaction. The bait-prey pairs can be sequenced individually, for example, from plasmids, PCR or rolling circle amplification (RCA) products, or from short sequence paired tags. The paired tags can be catenated into longer molecules and sequenced using conventional unidirectional or paired end sequencing methods to generate approximately a 6-10 tag pair sequences from each sequencing read. The linking of bait and prey tags allows thousands of baits to be screened against thousands of preys (e.g., complete libraries of both) simultaneously in a single transformation and screening experiment. The catenation of tag pairs allows approximately 10 or more pairs to be characterized for the cost of one hi-copy plasmid sequencing read (depending on the read length). By using a paired end sequencing approach, approximately 12-20 tag pairs can be gener...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com