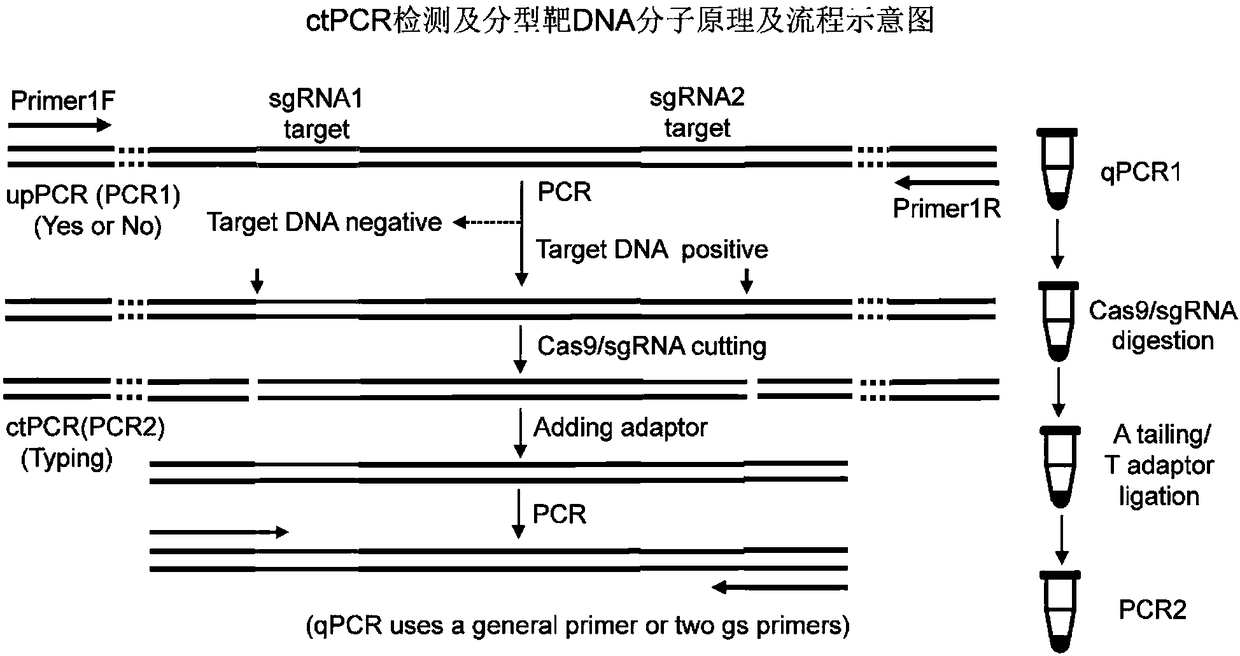
DNA detection and analysis method based on Cas9 nuclease and application thereof
An analytical method, nuclease technology, used in the field of biomedicine to achieve high specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
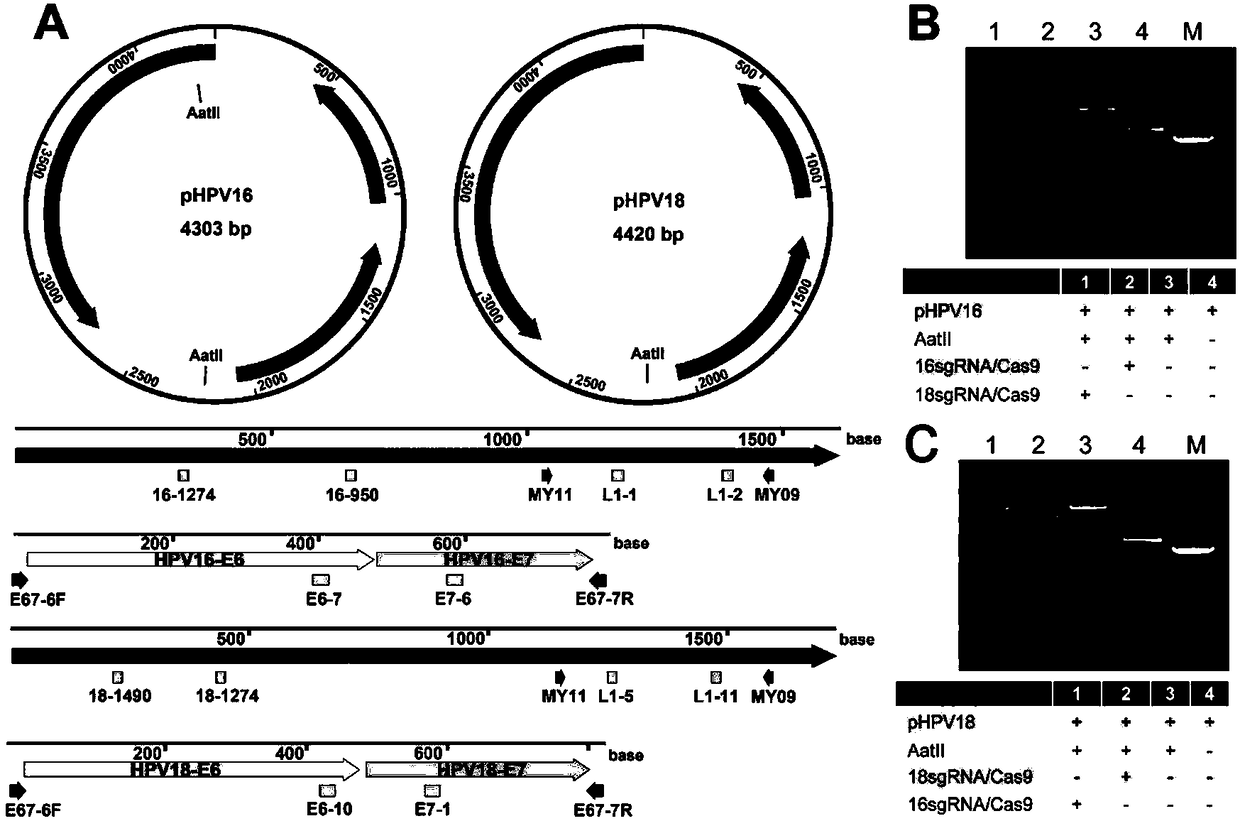
[0048] Example 1 Cutting HPV 16and 18L1 gene with Cas9 / sgRNA
[0049] experimental method:
[0050] Construction of sgRNA expression plasmid: first synthesize a pair of primers, use pCas9 (Addgene) as a template, and PCR amplify the prokaryotic Cas9 gene sequence, wherein the forward primer contains J23100 promoter plus RBS sequence (total 88bp). The PCR product was cloned into pCas9 plasmid with Cas9, trancRNA and spacer RNA sequences removed. The J23119-sgRNA sequence was amplified by PCR from pgRNA-bacteria (Addgene) and cloned into a freshly prepared pCas9 vector. The new plasmid was named pCas9-sgRNA, under the control of J23100 and J23119 promoters, pCas9-sgRNA can express Cas9 protein and sgRNA respectively. This plasmid also contains the chloramphenicol gene under the control of the cat promoter. Various sgRNA sequences cut by BsaI enzyme (annealed double-stranded oligonucleotides with BsaI sites at the ends) were cloned into pCas9-sgRNA, and Cas9 protein and sgrRNA...
Embodiment 2
[0055] Example 2 Cutting the HPV L1 gene cloned into the plasmid with Cas9 / sgRNA
[0056] experimental method:
[0057] Preparation of sgRNA: sgRNA was synthesized by in vitro transcription with T7 polymerase (New England Biolabs) according to the instructions. The sgRNA DNA template was amplified by three PCRs using the oligonucleotides listed in Table 1. The first PCR was performed with F1 and R (7 cycles). Use the product of the first PCR as a template, F2 and sgR as primers for the second PCR (30 cycles), use the product of the second PCR as a template, and F3 and sgR as primers for the third PCR (30 cycles) . The purified product of the third PCR was used as a template for in vitro transcription. The purified sgRNA template was then incubated overnight at 37°C with T7 RNA polymerase (NewEngland Biolabs) for in vitro transcription. The in vitro transcribed RNA was mixed with Trizol solution, extracted sequentially with chloroform and isopropanol, and precipitated with...
Embodiment 3
[0061] Embodiment 3 detects HPV 16 and 18L1 gene with ctPCR
[0062] experimental method:
[0063] Preparation of sgRNA: Same as Example 1.
[0064]Detection of HPV 16 and 18L1 genes cloned in plasmids by ctPCR: To prepare T-linkers, oligooJW102 and oJW103 (Table 3) were dissolved in Tris-HCl / EDTA / NaCl(TEN) buffer and mixed in the same molarity. The mixture was heated at 95 °C for 5 minutes and cooled slowly to room temperature. A pair of sgRNA specific to HPV16 and HPV18L1 genes combined with Cas9 protein cuts the plasmids of L1 gene clones of various HPV subtypes (200ng). The plasmid (200ng) was mixed with reaction buffer containing 1×Cas9 nuclease, 1μM Cas9 nuclease, 300nM sgRNA a (16-1274 or 18-1490; Table 2), 300nM sgRNA b (16-950 or 18-1274; Table 2 ) of the preassembled Cas9 / sgRNA complex and incubated at 37°C for 5 minutes. The digestion reaction (5 μL) was mixed with 5 μL premixed Taq (Takara) and incubated at 72 °C for 5 min plus A. Add A reaction solution (10 μ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com