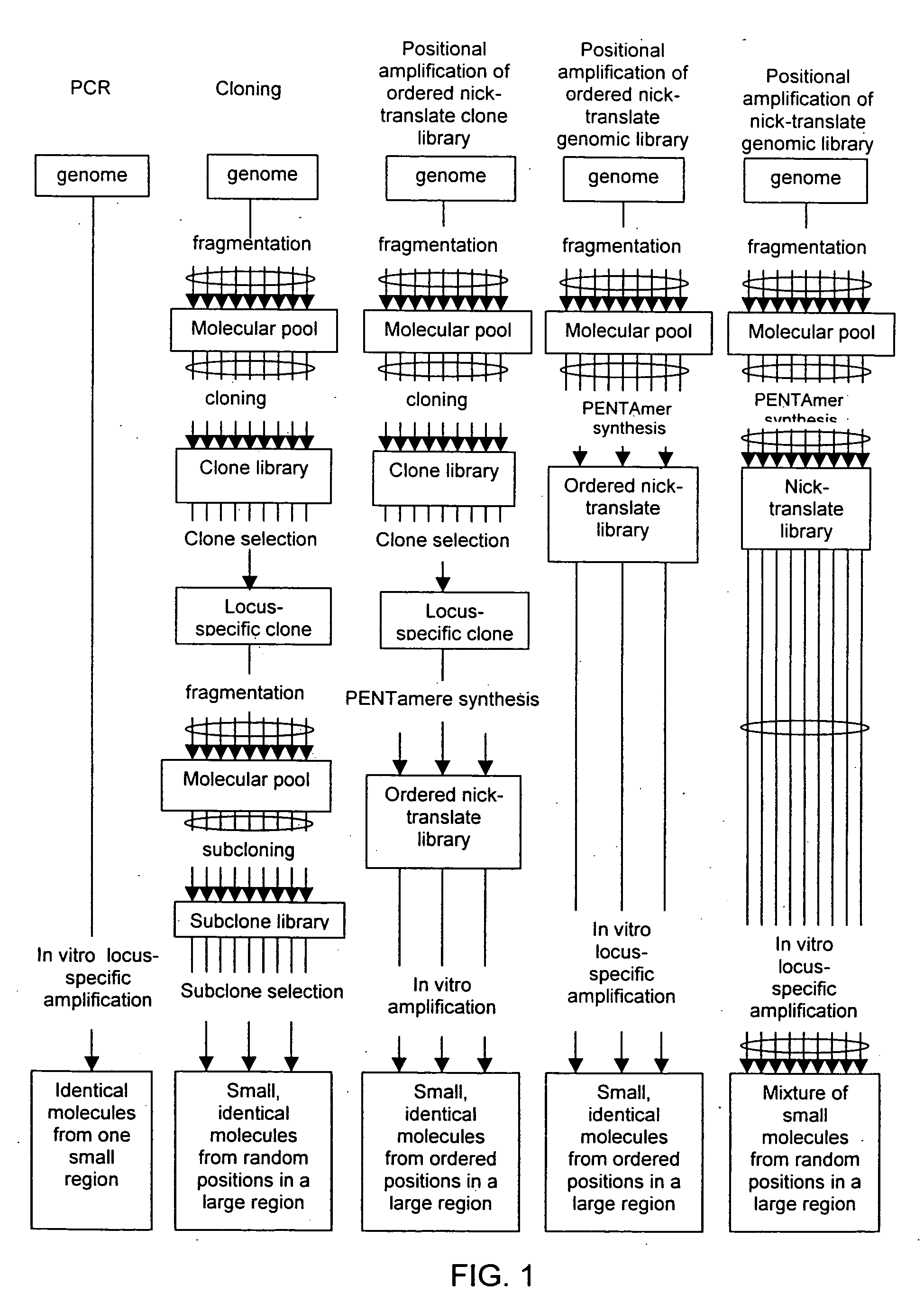
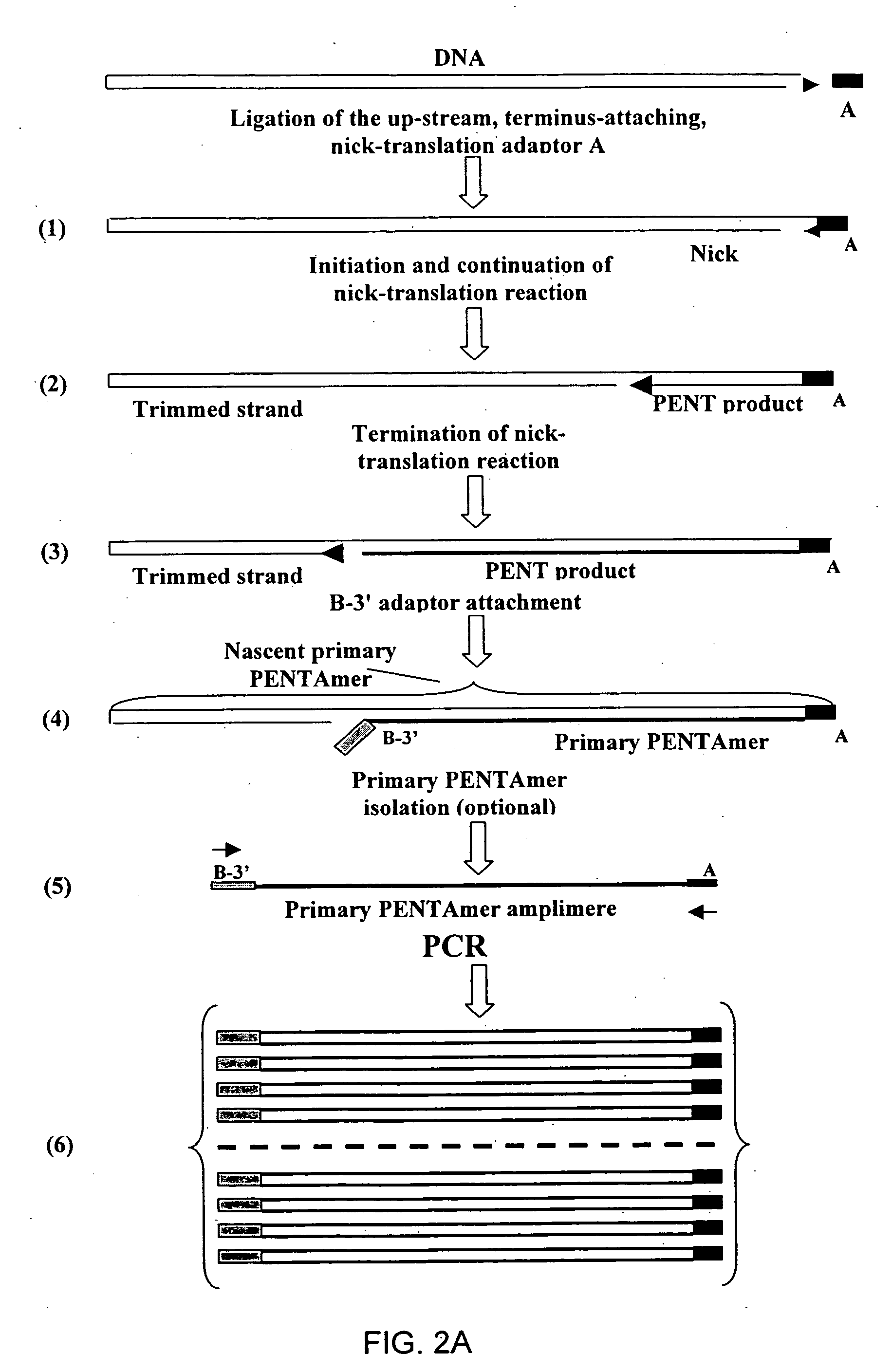
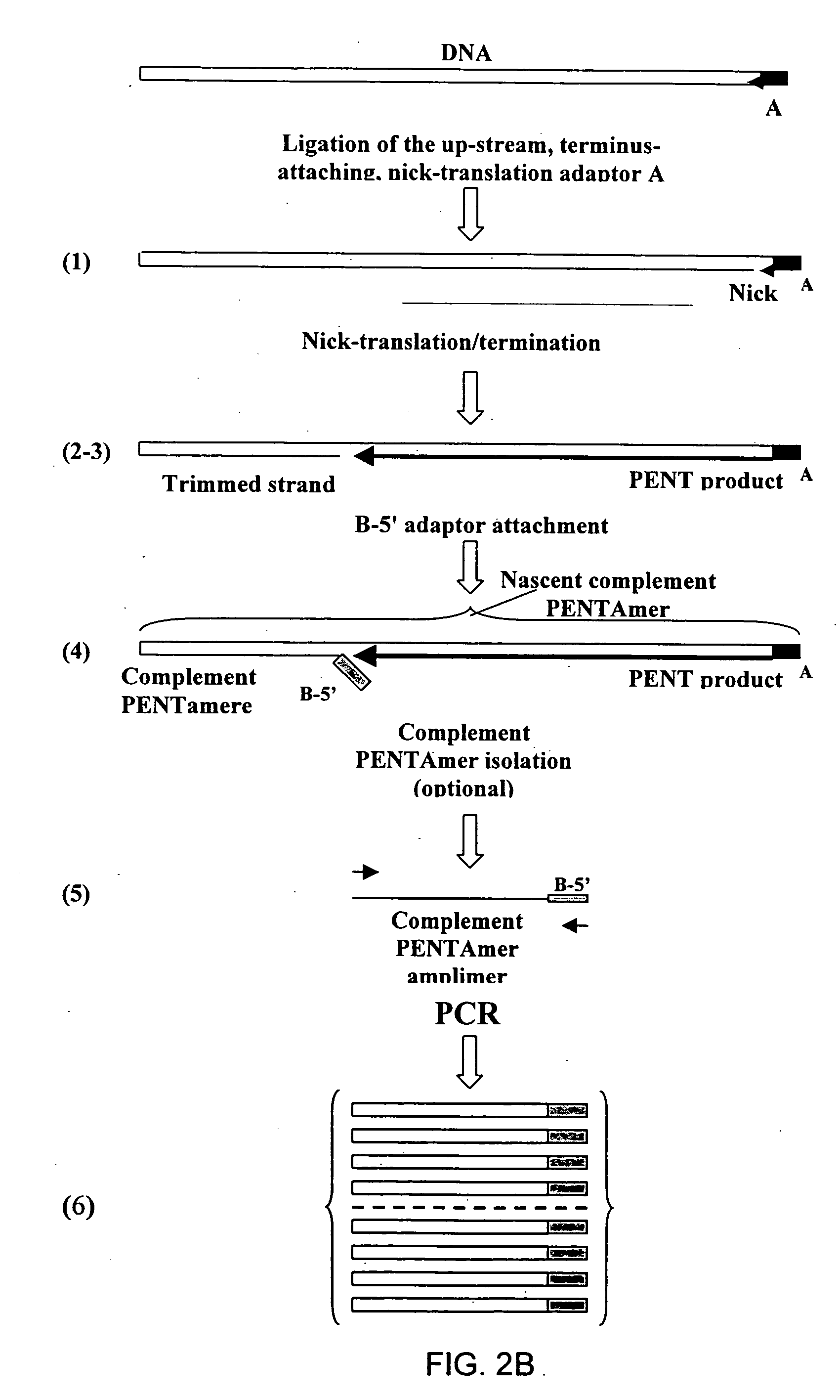
Method of producing a DNA library using positional amplification
a technology of positional amplification and dna library, applied in the field of molecular biology and biochemistry, to achieve the effect of fast and economical
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of PENT Adaptors.
[0904] This example describes the preparation of several types of adaptors used in different examples for terminal and internal tagging of the double-stranded DNA molecules. Oligonucleotide sequences are shown in Table 4.
[0905] Up-stream, terminus-attaching nick-translation adaptor A (FIG. 40) is prepared by annealing 100 pmol of oligonucleotide 5608 I and 100 pmol of the oligonucleotide 5602 I by cooling from 70° C. to room temperature at least 2 h in 20 μl of TE-0.1 (10 mM Tris-HCl pH 8.0, 0.1 mM EDTA). The annealed oligonucleotides are incubated with 5 U of Klenow enzyme (exo−) in 40 μl of 50 mM Tris-HCl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 50 μg / ml BSA, and in the presence of 100 mM dATP and 1 mM ddCTP at 37° C. for 1 h.
[0906] Acceptor-adaptor (AC) (FIG. 40) is prepared by dephosphorylation of 10 pmol of oligonucleotide 5608 I in 10 μl of 50 mM Tris-HCl, pH 8.5, 5 mM MgCl2 using 2 U of shrimp-alkaline phosphatase, SAP (Boehringer Mannheim; Indianapolis...
example 2
Efficient Ligation of Blocked PENT-Adaptors
[0913] Ligation of specialized nick-translation adaptors to the ends of DNA molecules is an important step towards the creation of a PENTAmer. This example describes the efficiency of ligation of a specialized 3′-end-blocked recombination nick-translation adaptor RA-(L-cos)(donor-adaptor Dn) with 5′phosphorylated 4-base GATC terminus to the recipient molecule (acceptor-adaptor AC) with complementary 5′ termini (Example 1).
[0914] Five reaction mixtures which contain 0, 200, 400, 800 and 800 nM adaptor RA-(L-cos) (donor Dn), 200 nM acceptor-adaptor (AC) in the first four tubes (no acceptor-adaptor in tube 5), 66 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM DTT, 1 mM ATP and 1 U of T4 DNA ligase (Boehringer Mannheim, Indianapolis, Ind.) in 10 μl are incubated for 2 h at 20° C. Tubes 6 and 7 contain ligase-deficient controls with 200 nM adaptor-acceptor and 800 nM adaptor-acceptor, respectively. The products of the ligation reactions are analyzed on...
example 3
Preparation of the “PENT-Ready” Lambda DNA Bam HI templates.
[0916] This example describes the preparation of lambda DNA / Bam HI restriction fragments with upstream nick-translation adaptors A, which are used in Examples 4-7, and 9-14.
[0917] Following the incubation of 5 μg of lambda DNA with 20 U Bam HI (Boehringer Mannheim, Indianapolis, Ind.) in 25 μl of 10 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 100 mM NaCl, 1 mM 2-mercaptoethanol for 2 h at 37° C., the mixture is supplemented with 3 μl of shrimp alkaline phosphatase (SAP) buffer (Boehringer Mannheimn) and 2 U of SAP (Boehringer Mannheim), and incubated for 30 min at 37° C. After heat inactivation of SAP at 68° C. for 15 min the DNA is precipitated with ethanol, washed with 70% ethanol, dried and dissolved in 31 μl TE (10 mM Tris-HCl pH 8.0, 1 mM EDTA) with a final molar concentration of Bam HI ends equal to 50 nM. Then, 5 μl of SAP treated Barn HI lambda DNA restriction fragments (250 fmol ends) are ligated with 1 pmol of nick-transla...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| ionic strength | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com