A method and system for quickly comparing gene data
A genetic data and comparison technology, applied in the field of genetic analysis, can solve the problems of low comparison efficiency in the genetic data analysis process, complex algorithm structure, and long time-consuming, so as to improve throughput performance, reduce computing resources, and improve efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
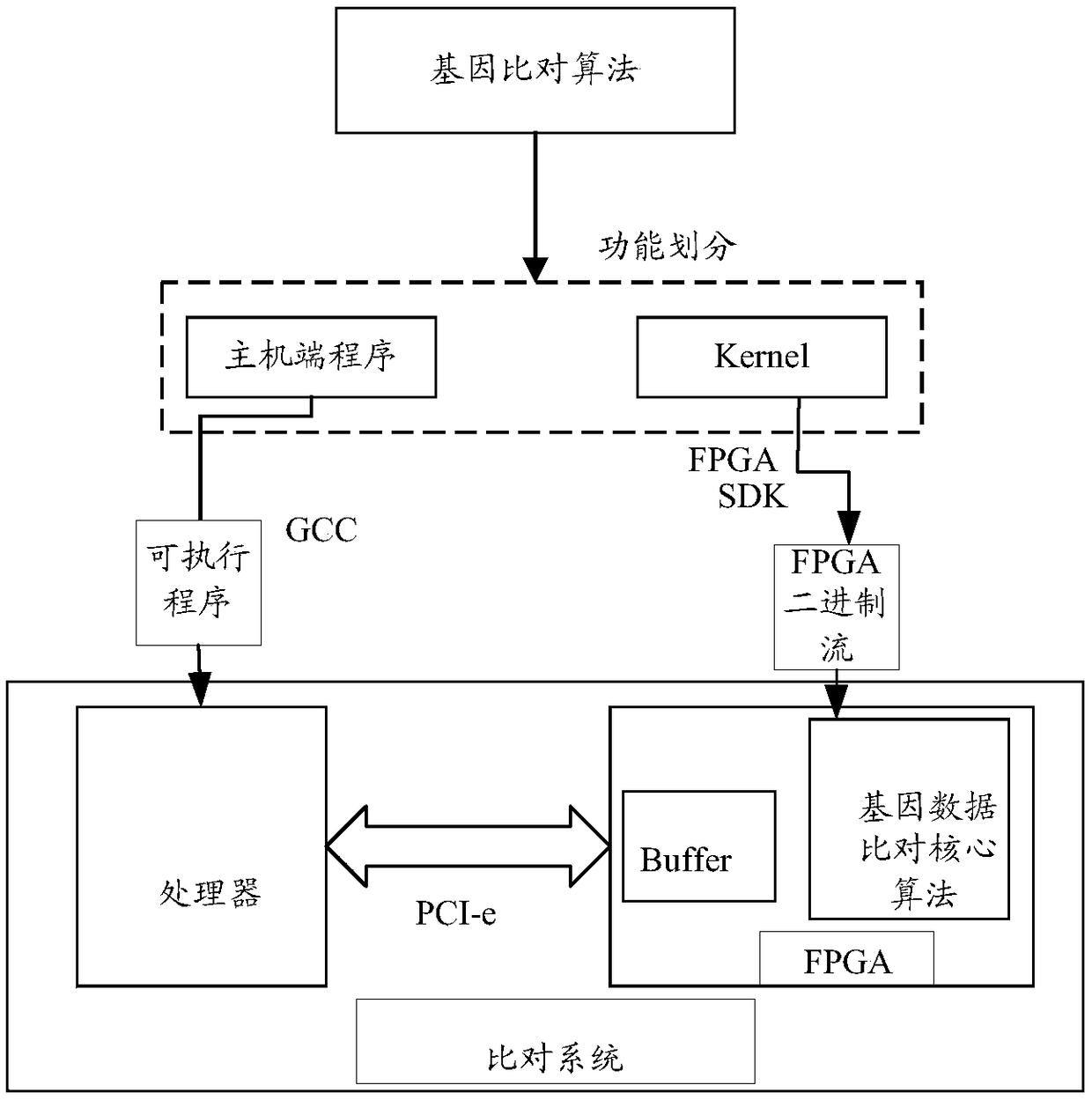
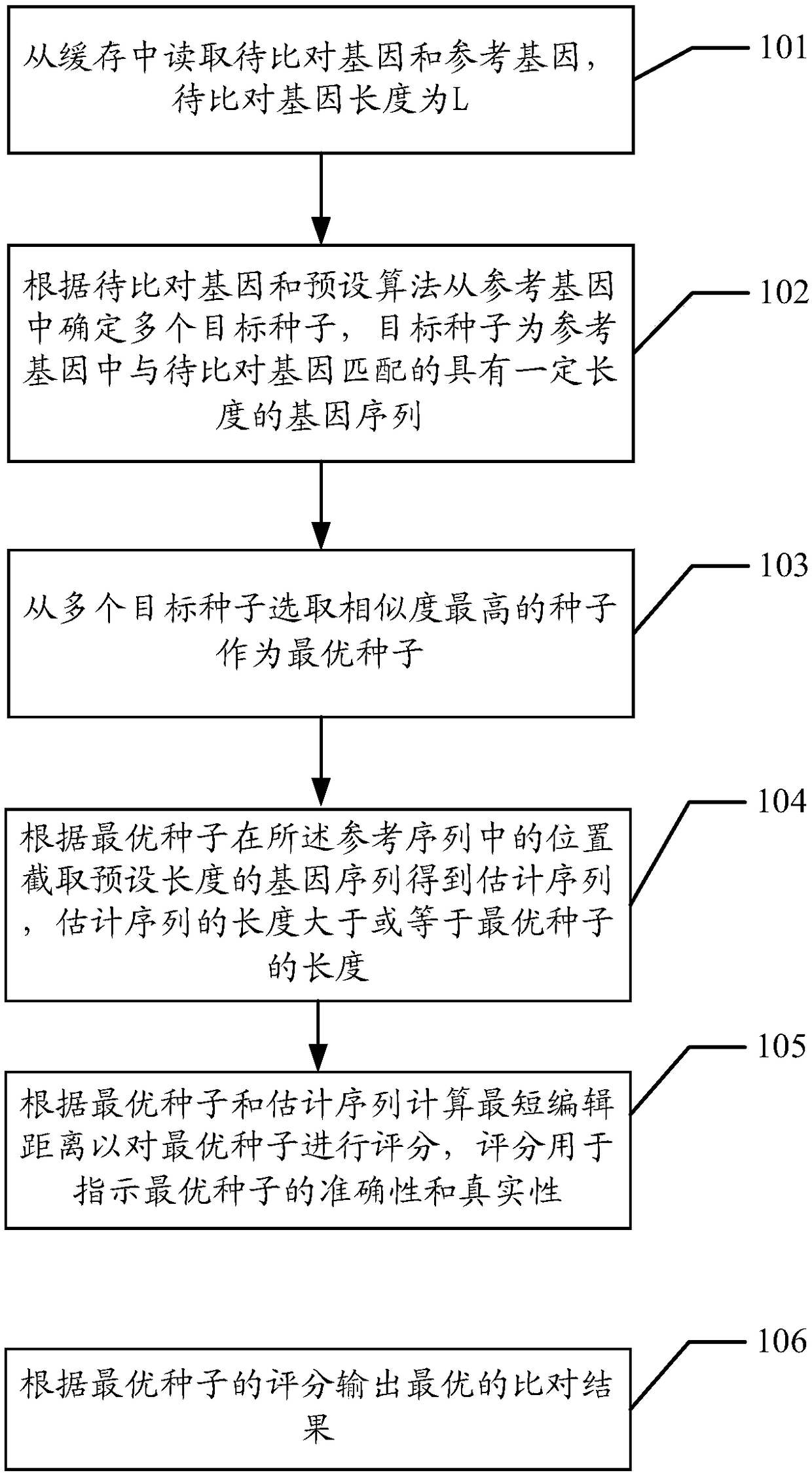
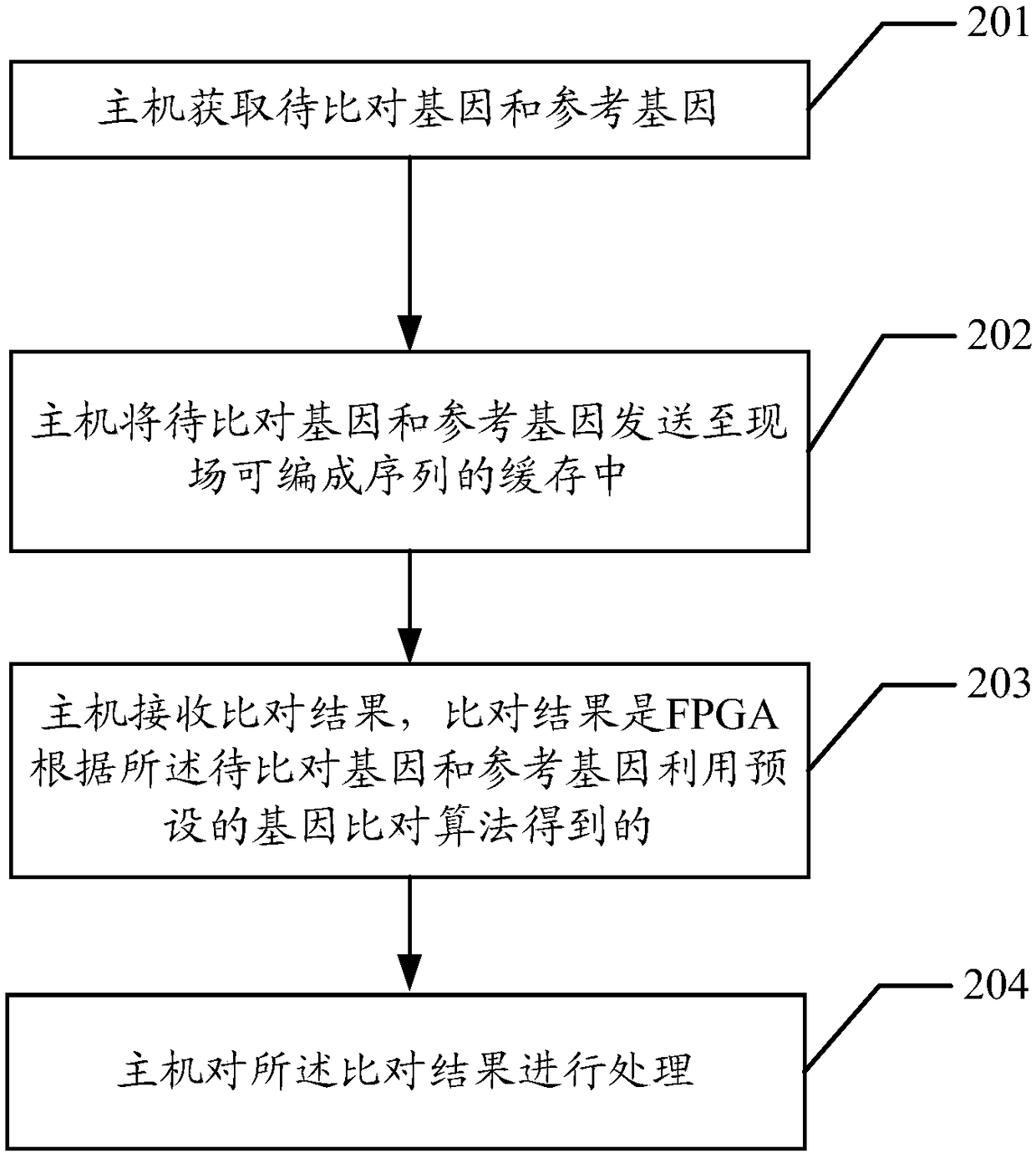
[0067] The embodiment of the present application provides a method for quickly comparing genetic data. By mining the parallelism and pipeline process in the comparison process, the host is used in combination with the FPGA. The host is responsible for preprocessing the genes to be compared and the reference genes. The FPGA hardware platform executes the core module of the gene data comparison algorithm, which can improve the parallelism of the algorithm through the optimization of parallel pipeline, thereby improving the throughput performance of the algorithm execution, improving the efficiency of gene comparison, and reasonably integrating the obtained seeds. Reducing computing resources and optimizing from various aspects can achieve the effect of accelerating the gene comparison process.
[0068] The terms "first", "second", "third", "fourth", etc. (if any) in the description and claims of the present invention and the above drawings are used to distinguish similar objects,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com