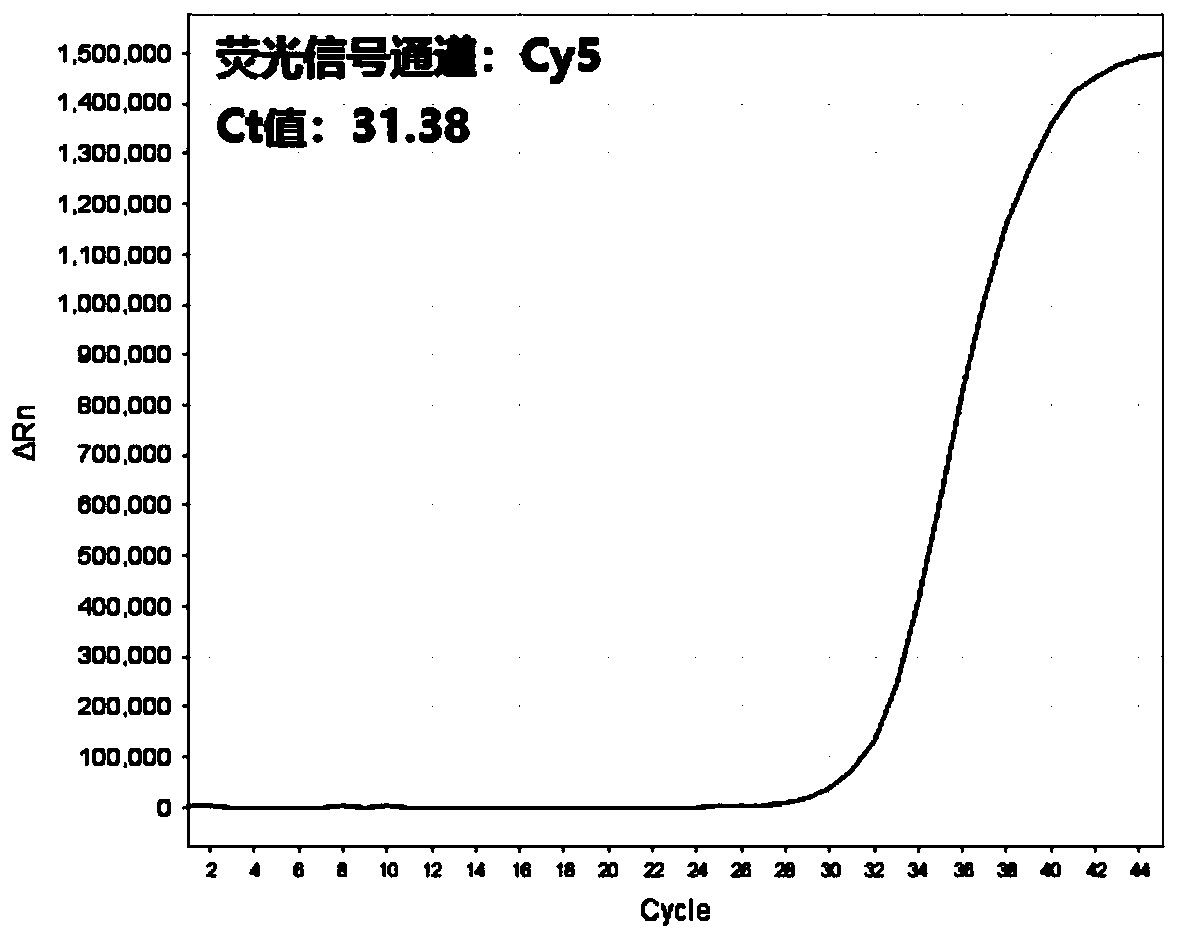
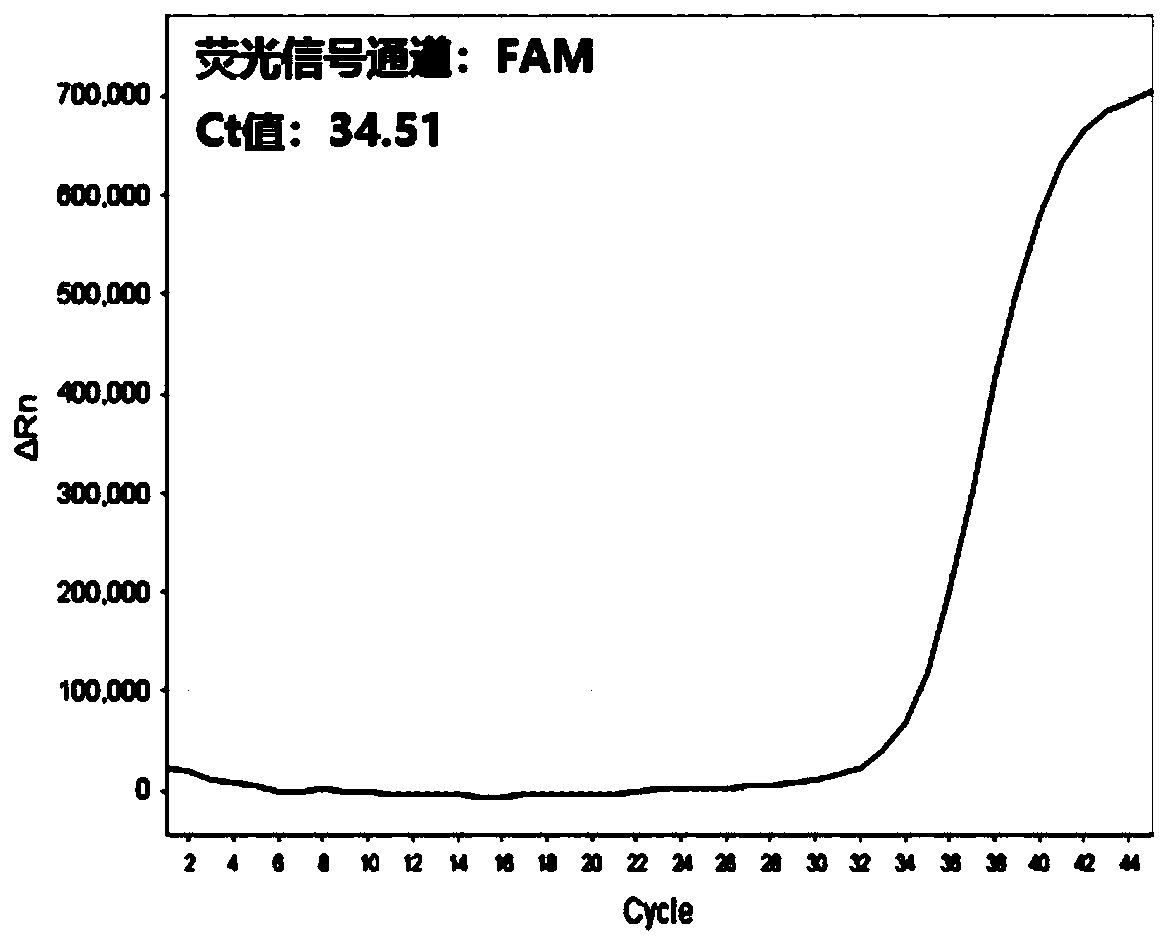
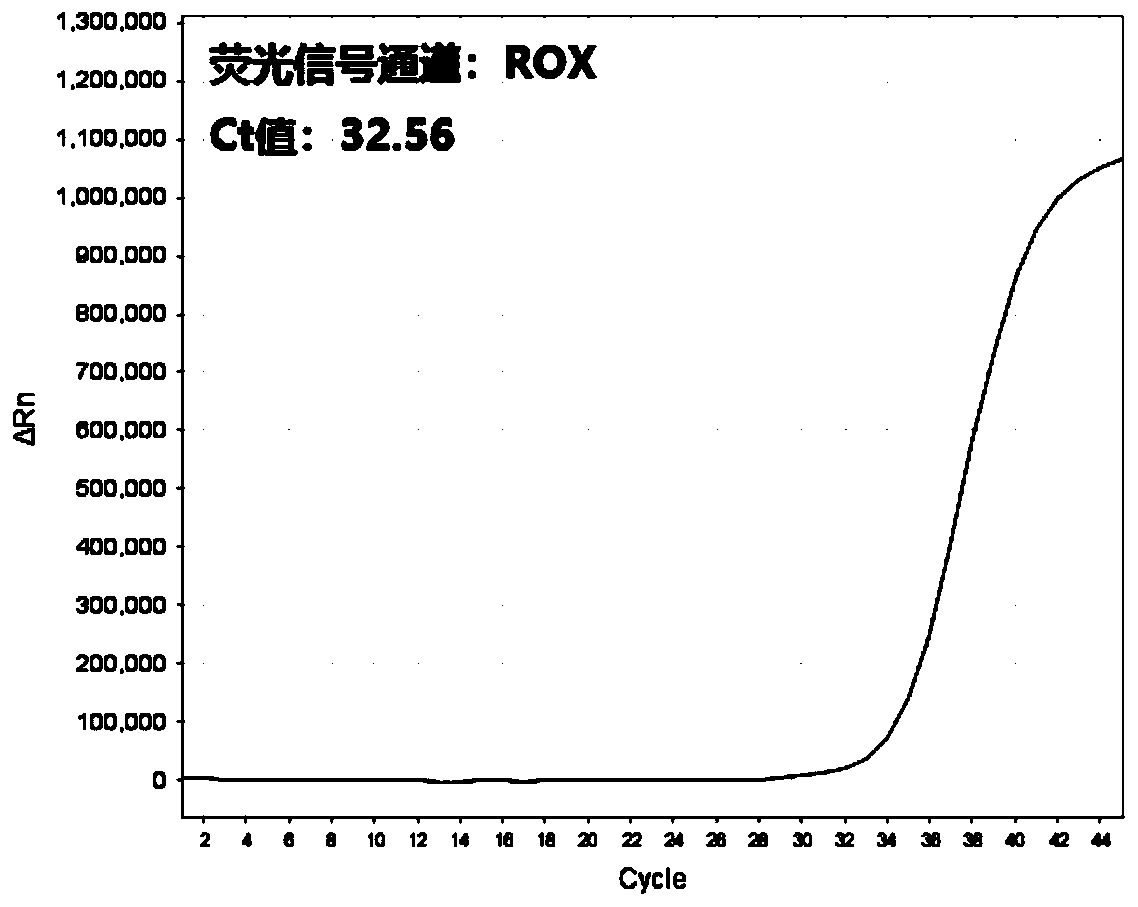
Multi-real-time fluorescent quantitative PCR kit, method and primer probe composition for detecting 2019 novel coronavirus
A real-time fluorescence quantitative and coronavirus technology, applied in the field of nucleic acid detection, can solve problems such as dyspnea, death, pneumonia, etc., and achieve the effects of short detection cycle, high sensitivity, and low missed detection rate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] The presence of 2019-nCoV RNA in the respiratory and blood samples of suspected patients and non-suspected patients was detected by multiple reverse transcription real-time fluorescent quantitative PCR detection kits for 2019-nCoV respectively (respiratory tract samples of suspected patients are numbered A1, blood samples of suspected patients The number is A2, the number of respiratory samples from non-suspect patients is B1, and the number of blood samples from non-suspect patients is B2):
[0054] 1. Nucleic acid extraction: Use nucleic acid extraction reagents to automatically extract nucleic acids from samples A1, A2, B1, and B2 on an automatic nucleic acid extraction instrument at the same time, and store the extracted nucleic acid samples at -20°C.
[0055] 2. Prepare RT-PCR system: Prepare RT-PCR reaction system according to 14 μL 2019 novel coronavirus detection master mix, 1 μL RT-PCR enzyme solution and 5 μL nucleic acid sample to be detected for each reaction...
Embodiment 2
[0059] 2019 Novel Coronavirus Multiplex Reverse Transcription Real-Time Fluorescent Quantitative PCR Detection Kit Standard Curve Establishment and Sensitivity Test:
[0060] 1. The positive recombinant plasmids containing S gene, ORF1ab gene, N gene and B2M gene were respectively diluted 10 times after the concentration and purity were determined, and obtained from 1 × 10 2 ~1×10 8 A total of 7 dilutions of positive plasmids in copies / μL were used as standard templates. Prepare the reaction system according to Table 4, perform multiple reverse transcription real-time fluorescent quantitative PCR according to Table 5, obtain the fluorescence amplification curve and draw the standard curve.
[0061] 2. For the S gene ( Figure 17 ), at a concentration of 1×10 2 ~1×10 8 The amplification curve within the range of copies / μL presents a typical S-shape, the curves are evenly spaced, and have good correlation. The linear equation is y=-2.9351x+41.435, R 2 =0.9981, the lowest de...
Embodiment 3
[0063] Multiple reverse transcription real-time fluorescent quantitative PCR detection kit for 2019 novel coronavirus Specificity test:
[0064] 1. Using pathogenic microorganisms of common respiratory diseases (including human coronavirus 229E, human coronavirus NL63, human coronavirus OC43, human coronavirus HKU1, influenza A virus, parainfluenza virus, metapneumovirus, influenza B virus, respiratory Syncytial virus, rhinovirus, boca virus, Chlamydia pneumoniae, Chlamydia trachomatis, Mycoplasma pneumoniae, and adenovirus) were used as templates to test the specificity of the kit.
[0065] 2. The test results are shown in Table 6. The results showed that the multiplex reverse transcription real-time fluorescent quantitative PCR detection kit for 2019 novel coronavirus was negative for pathogenic microorganisms of common respiratory diseases, which proved that the kit had good specificity.
[0066] Table 6 Kit specificity verification
[0067]
[0068]
[0069] The pres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com