Method for single cell classification and screening and device therefor
A classification method and technology of a classification device, applied in the field of bioinformatics, can solve the problems of cumbersome preparation process, lack of accuracy evaluation, low sensitivity of subgroup classification, screening and detection, etc., and achieve the effect of improving accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0134] Example 1: Classification of renal cancer single cells
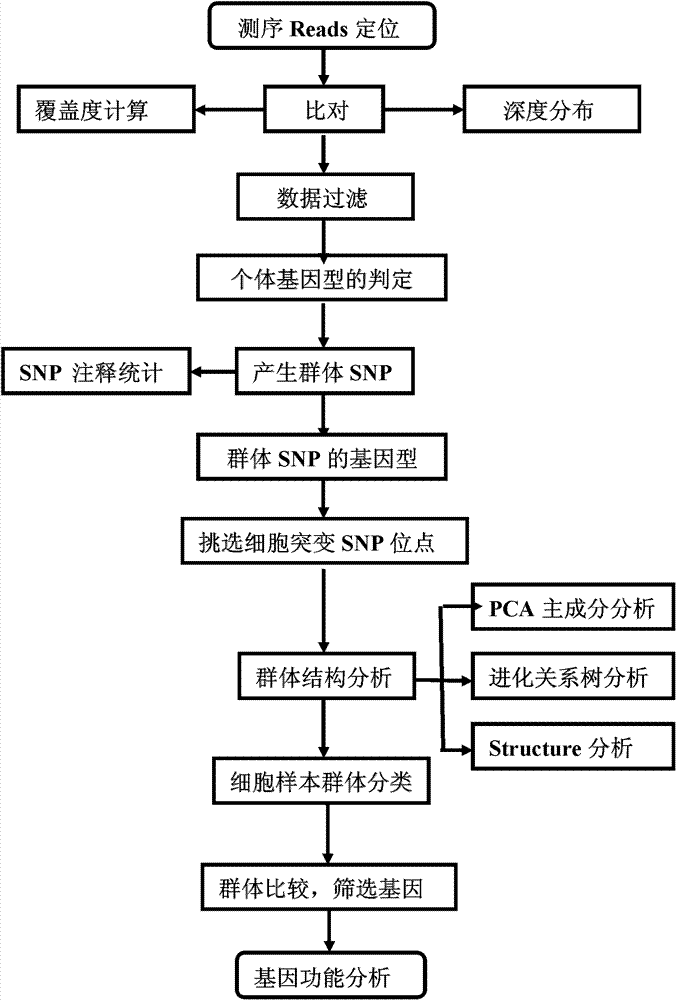
[0135] 1-1. Reads positioning
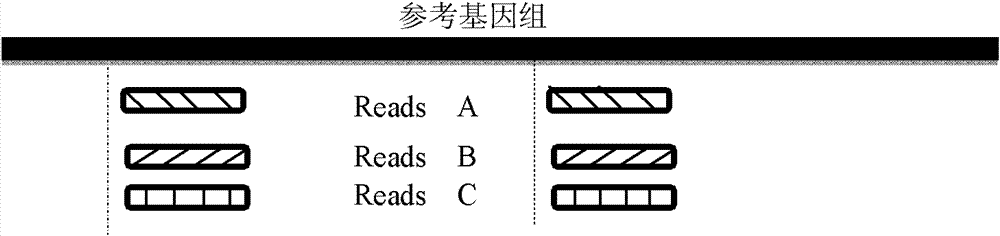
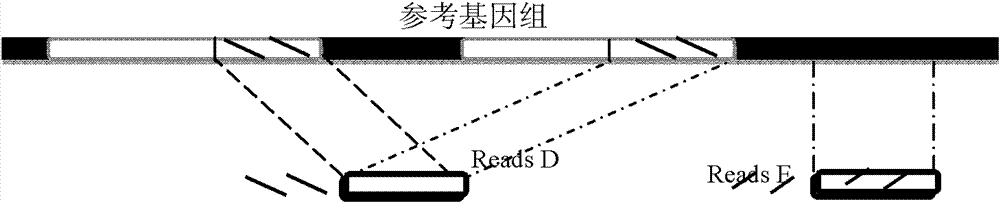
[0136] The reads results obtained by sequencing each single-cell sample were compared with the reference genome sequence (human genome HG18) using SOAPaligner alignment software (http: / / soap.genomics.org.cn / soapaligner.html). Two thousand copies and the read length of Reads is 100bp, so when SOAP is compared, each Reads is set to have a maximum of 3 misalignments (Mismacth), and Gap parameters are not allowed to ensure that the positions of Reads that can be compared are accurate.
[0137] 1-2. Basic data statistics
[0138] According to the above comparison results, the sequencing depth and coverage of each sample (single cell or tissue) relative to the reference genome sequence are calculated. After statistics, when the whole genome is sequenced and the Mean Depth is around 3×, due to PCR amplification There is a certain bias (Bias), so the coverage of the sample fluctuates grea...
Embodiment 2
[0229] Example 2: Classification and screening of leukemia single cells
[0230] 2-1. Reads positioning
[0231] A 30× depth exome sequencing was performed on each single cancer cell, and the obtained reads results were compared with the reference genome sequence (human genome HG18) using SOAPaligner2.0 alignment software. Since the number of human SNPs is 2 / 1000 and the read length of Reads is about 100bp, we set each Reads to have a maximum of 2 mismatches (mismacth) during SOAP alignment, and Gap is not allowed to ensure that the alignment is to the reference The accuracy of Reads on the genome.
[0232] 2-2. Basic data statistics
[0233] A total of 53 cancer cells and 8 oral epithelial cells (normal cells) were sequenced. Table 5 shows the coverage and depth numerical information of exome sequencing for each cell sample.
[0234] Table 5 Coverage and depth of exome sequencing for each cell sample
[0235]
[0236]
[0237] 2-3. Data filtering
[0238] Same as ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com