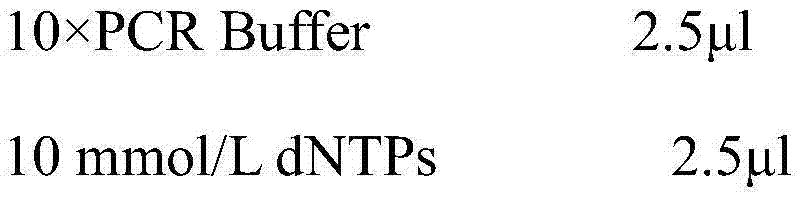Molecular specific marker primers for No. 4 and No.32 of an improved variety Changlin of Camellia oleifera and an identification method
A technology for labeling primers and Camellia oleifera seeds, which can be used in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc. It can solve the problems of long cycle, poor repeatability and low specificity, and achieve the effect of simple method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
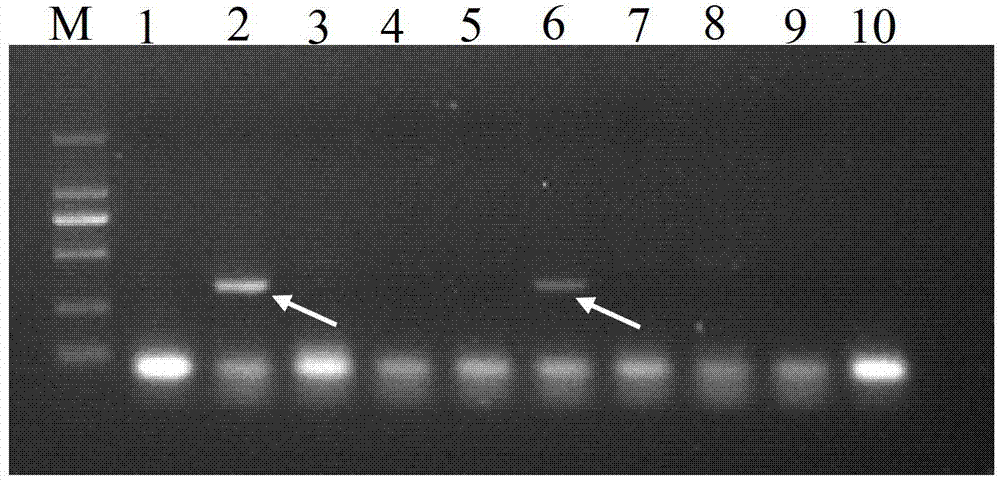
[0036] (1) Extraction of genomic DNA of Camellia oleifera:
[0037] Take 0.01 g of the young leaves of the improved Camellia oleifera to be tested, add liquid nitrogen and grind them thoroughly, and use the SDS-CTAB method to extract the genomic DNA, and extract the crude genomic DNA of the improved Camellia oleifera through multiple extractions. The crude DNA was purified by Magabio Nucleic Acid Purification Kit (Bioer, Hangzhou, China), and detected by 1.5% agarose gel electrophoresis and DNA / RNA UV spectrophotometer (GeneQuant Pro, GE Healthcare). Integrity, Purity and Concentration. OD 260 / OD 280 DNA samples >1.8 were used for subsequent PCR amplification. DNA extracts were stored in a -20°C refrigerator for later use.
[0038] (2) Design specific PCR amplification primers, the sequence of the primer pair is:
[0039] The upstream primer 5'-CCTATGGTGCCTCTTTGTTTGATGT-3' and the downstream primer 5'-GTGACAAGTTGAACTCTAGCACAAA-3' were synthesized by Shanghai Bioengineeri...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com