Molecule specificity marker primer and identification method of improved variety of camellia oleifera Changlin Number 55
A technology for marking primers and improved varieties of camellia oleifera, applied in the direction of DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of long cycle, complicated PCR amplification map, and difficult variety identification, and achieve the effect of simple method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
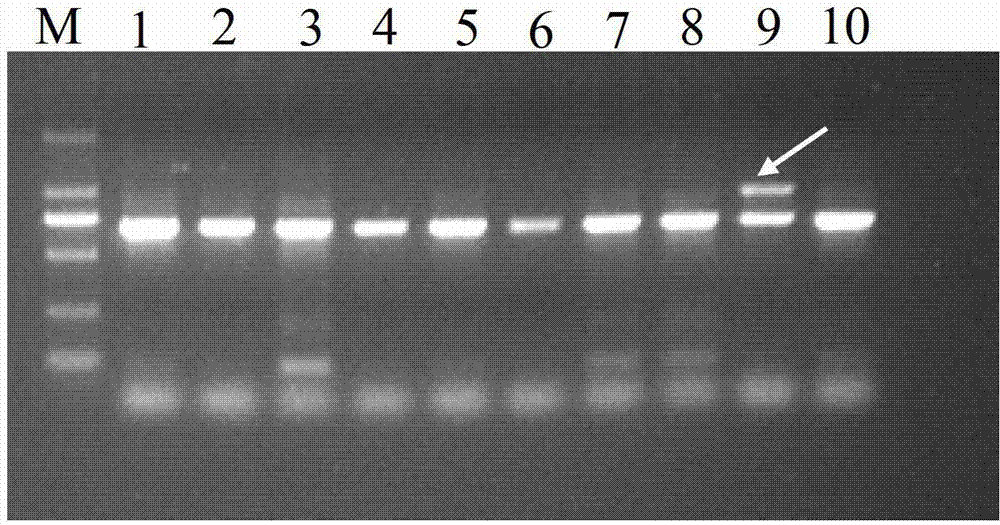
[0045] (1) Extraction of genomic DNA of improved Camellia oleifera: Take 0.01g of young leaves of Camellia oleifera to be tested, add liquid nitrogen to grind thoroughly, and use SDS-CTAB method to extract genomic DNA. Genomic DNA crude extract. The crude DNA was purified by Magabio Nucleic Acid Purification Kit (Bioer, Hangzhou, China), and detected by 1.5% agarose gel electrophoresis and DNA / RNA UV spectrophotometer (GeneQuant Pro, GE Healthcare). Integrity, Purity and Concentration. OD 260 / OD 280 DNA samples >1.8 were used for subsequent PCR amplification. DNA extracts were stored in a -20°C refrigerator for later use.
[0046] (2) Design specific PCR amplification primers. The sequence of the primer pair is the upstream primer 5′-CATACATACACTTCCTAAGCCAAAA-3′ and the downstream primer 5′-GTTCAAGCATTGTTCAAGCACTC-3′, which were synthesized by Shanghai Bioengineering Technology Co., Ltd.
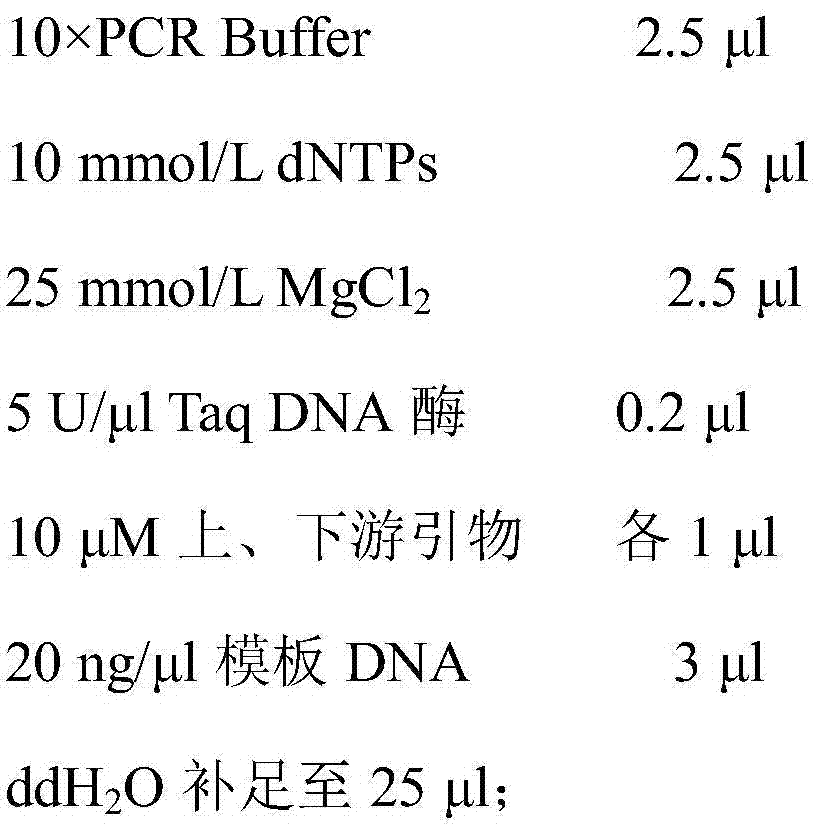
[0047] (3) PCR amplification of SCAR molecular markers:
[0048] Composition of P...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com