Soybean chalcone reductase gene chr3 and its application
A reductase and gene technology, applied in the field of biosynthesis of plant secondary metabolite daidzein, can solve the problems related to unclear synthetic ability and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Example 1 Cloning of Soybean Chalcone Reductase Gene CHR3
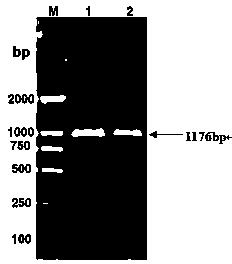
[0021] The soybean variety "Jinong 17" (seeds provided by the Biotechnology Center of Jilin Agricultural University) was selected and planted in the field. When the plants grew to the flowering stage, the leaf tissue was selected, and the total RNA was extracted using the kit RNAiso Plus, and followed by the American Fermentas company. Reverse transcription was performed according to the instructions of the reverse transcription kit, and the first strand of cDNA was synthesized. Primers P1 and P2 were designed according to an incomplete gene fragment of unknown function (No. TC227002) contained in the soybean EST library of the National Center for Biological Information (NCBI). RT-PCR was used to amplify specific fragments using cDNA as a template. Then, the RACE kit from TAKARA Company was used to clone the 3' end fragment and the 5' end fragment of the target gene respectively. After the sequences at both...
Embodiment 2
[0027] Example 2 Construction of Plant Expression Vector PBI121-CHR3 Gene
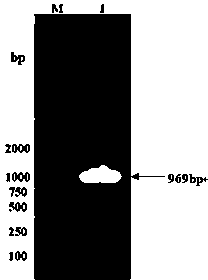
[0028]The gene sequence of the open reading frame was predicted by the special analysis software on the website of the National Center for Biological Information (NCBI), and the predicted result was a fragment with a length of 969 bp, and the open reading frame was at 1-969 bp. According to the cDNA sequence of soybean chalcone reductase CHR3 gene, design primers with restriction enzyme sites and amplify the complete coding reading frame, add XbaⅠ enzyme site at the upstream, add BamHI enzyme site at the downstream, and add a BamHI enzyme site at the upstream Primer: GGG TCTAGA ATGGCAGGAAAGAAAATC, downstream primer: TTT GGATCC TCAAACGTCTCCATCCC, using the cDNA obtained in Example 1 as a template for PCR amplification, and connecting the amplified fragment of CHR3 with upstream and downstream restriction sites to the PMD-18T vector . Then digest the plasmid vector of the CHR3 gene with XbaⅠ / BamHI coh...
Embodiment 3
[0030] Example 3 The cultivation of transformed CHR3 gene tobacco
[0031] Then the recombinant expression vector pBI121-CHR3 was transformed into competent Agrobacterium DH105 by heat shock method. Tobacco explants were prepared for transformation with Agrobacterium. Select tobacco seeds with full grains, sterilize them with 70% alcohol and 5% sodium hypochlorite solution, wash them with sterile deionized water, inoculate them on the tobacco germination medium, and cultivate them in a bright place for 15 days; transform Agrobacterium in YEP The liquid medium was cultured with shaking at 28°C until the OD600 value was 0.6-0.8, and the bacteria were collected by centrifugation and resuspended with MS liquid medium. Cut off the sprouted tobacco 1cm 2 Leaves of the same size and punched a number of holes, put them into the MS medium containing transformed Agrobacterium and suspended them for 8 minutes, then placed the leaves on the common medium of tobacco, and cultivated the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com