A False Positive Gene Mutation Filtering Method for Targeted Capture Genetic Sequencing Data
A technology of gene sequencing and target capture, applied in the field of data science, can solve the problems of small target area, difficult to obtain training data, unable to cope with batch differences, etc., and achieve the effect of small scale
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
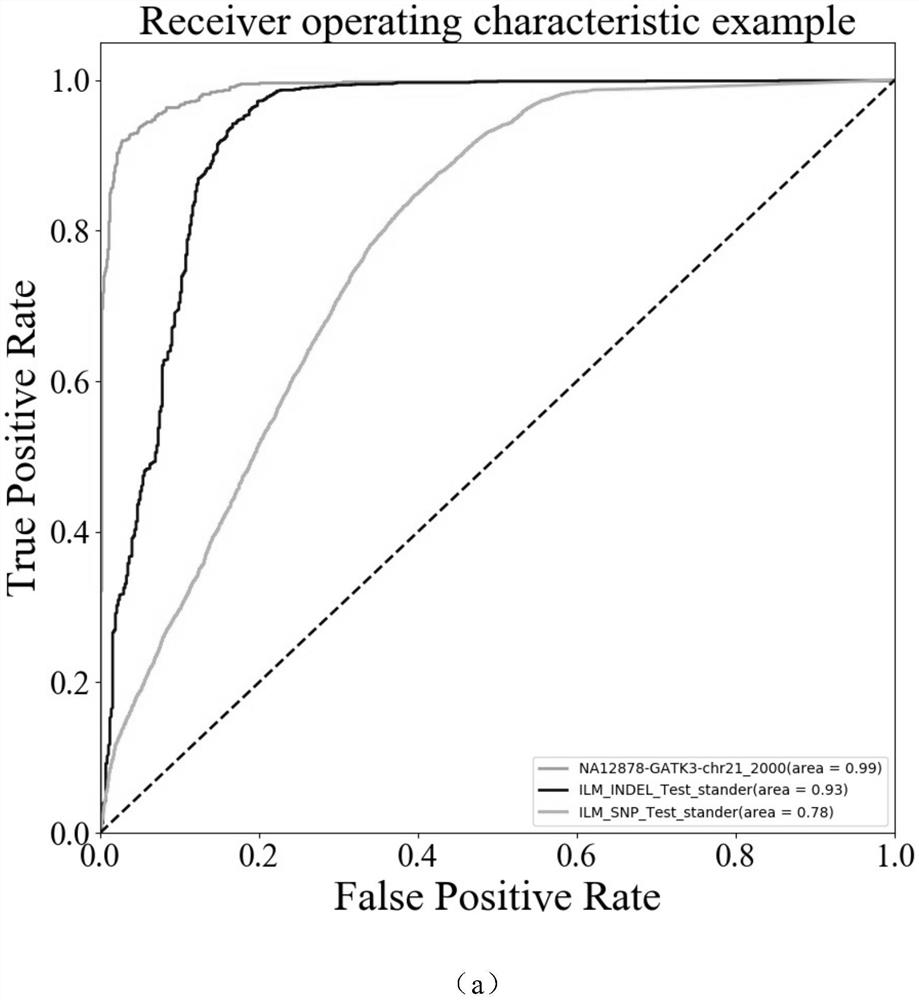
[0051] see Figure 4 , a method for filtering false positive gene mutations aimed at targeted capture gene sequencing data of the present invention, comprising the following steps:
[0052] S1. Preprocessing the sample data of genetic detection mutation sites;
[0053] S101. Read the gene mutation detection report file output by the mutation detection software, that is, the output file conforming to the VCF format standard, read the data of each attribute from it, and use data standardization and normalization methods to preprocess the sample data. In the data preprocessing module, the feature data and descriptions of the mutations extracted from the VCF format file are shown in Table 1.
[0054] Table 1 The characteristics and physical meaning of mutations extracted from VCF format files
[0055]
[0056]
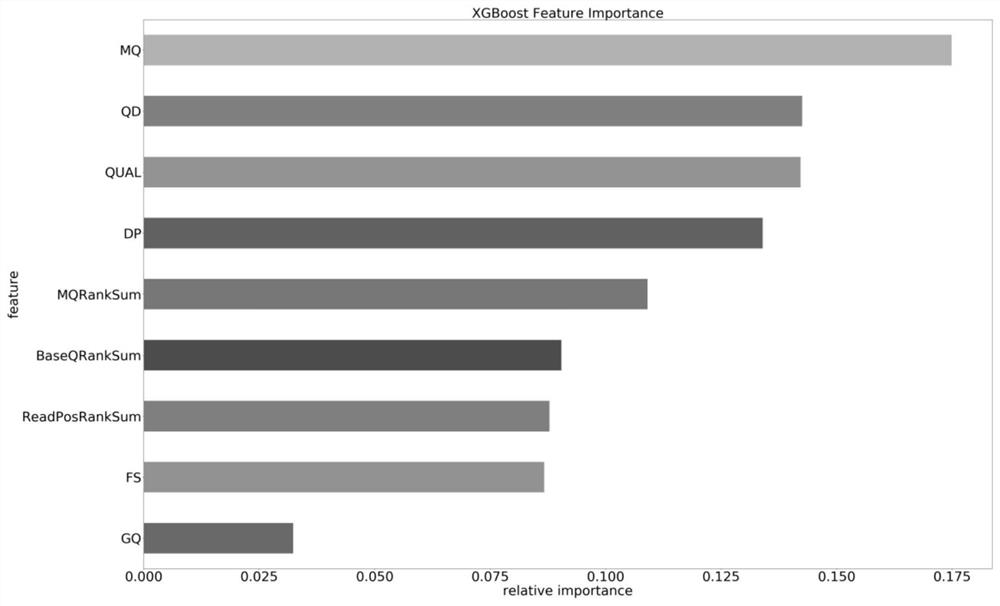
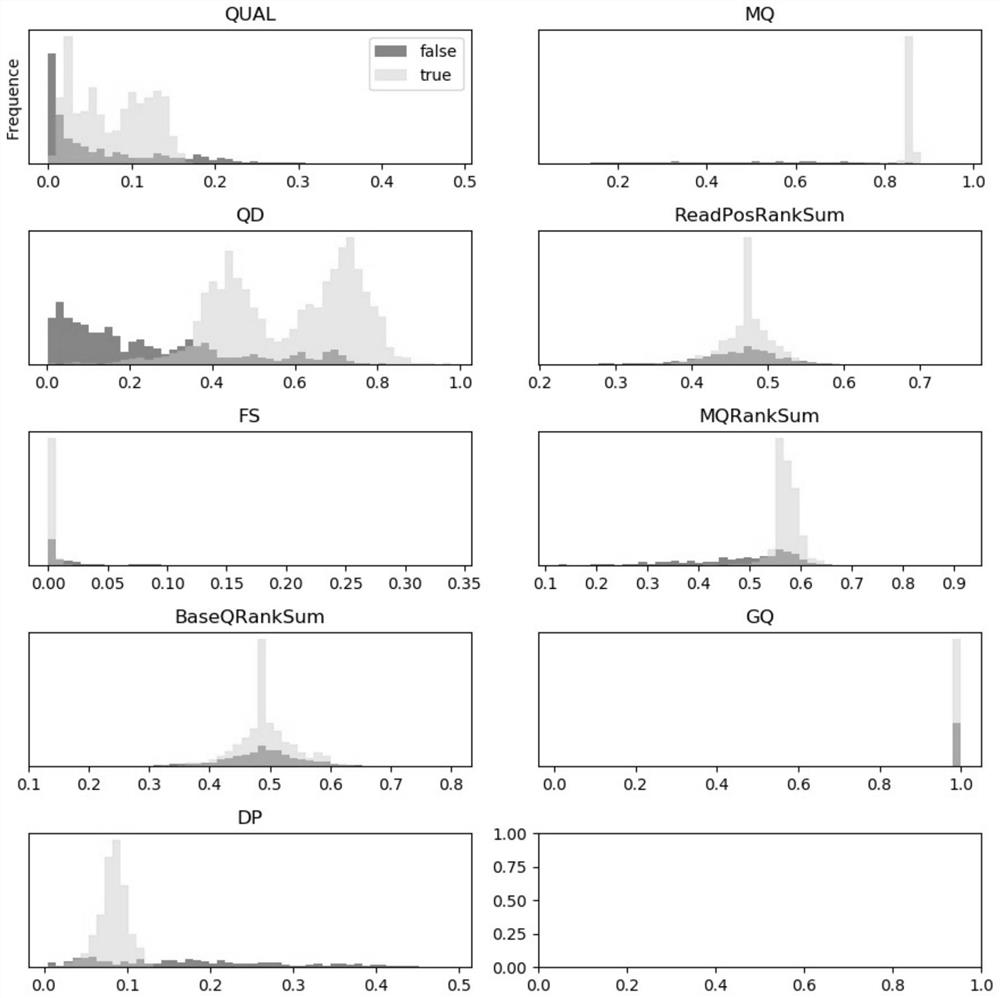
[0057] In order to ensure the relevance and importance of the selected features, the feature selection engineering method in machine learning is adopted, and the t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com