SNP marker combinations for inferring major ethnic groups in northwestern China and adjoining Central Asian countries
A national and labeling technology, applied in the biological field, can solve problems such as short amplification fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
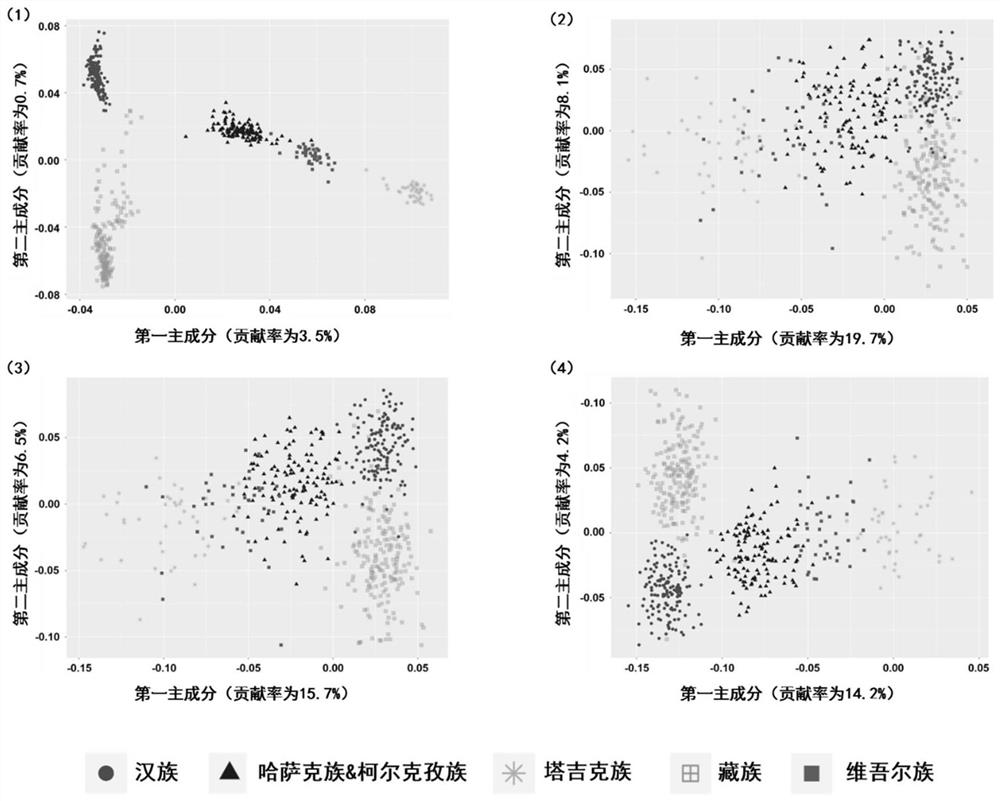
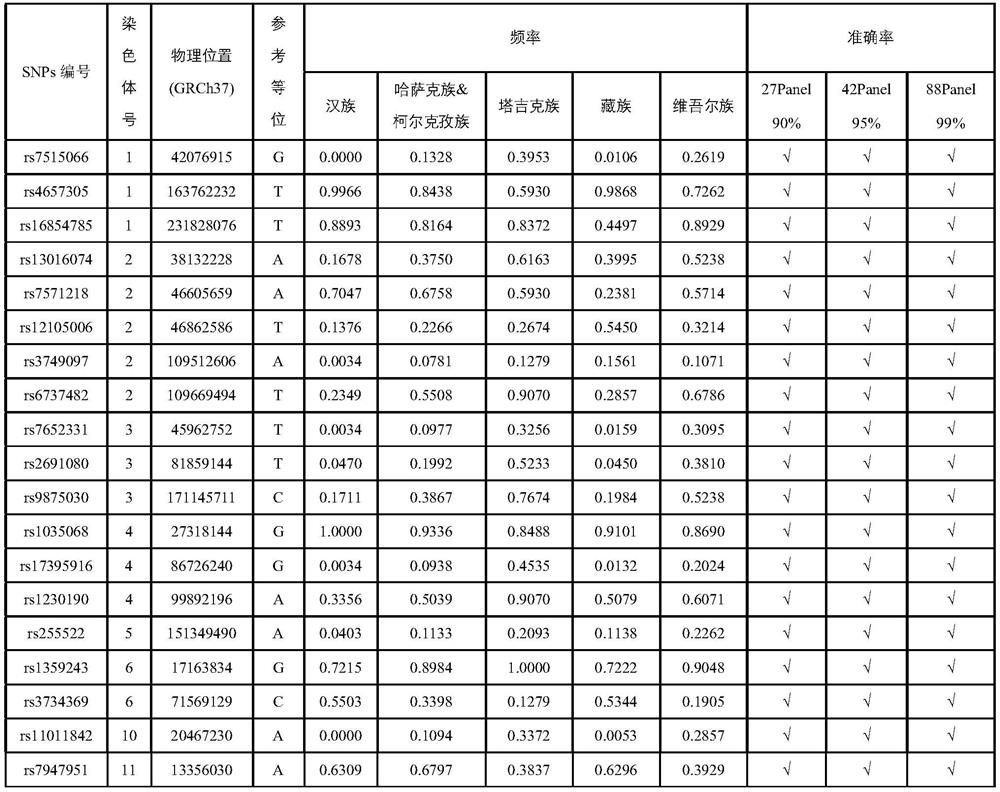
[0039]This embodiment is based on the article Characterizing private and shared signatures of positive selection in 37Asian populations published by Xuanyao Liu, et al. in European Journal of Human Genetics (2017) and Genomic by Jinchuan Xing, et al. published in PLOS Genetics (2013). Analysis of natural selection and phenotypic variation in high-altitude Mongolians article data screened out six geographical populations of Han, Tibetan, Kazakh, Kirgiz, Uyghur and Tajik, combined with self-collected samples, a total of 551 samples of genomic level SNP data. The population and sample size included in each region are shown in Table 2. After the integration, merging, site quality control and individual screening of data from different sources, a raw data set including 551 unrelated individuals, each containing 150,793 SNP sites, was formed for subsequent ancestral indicative SNPs Construction of marker combinations.
[0040] Table 2. Sample sources
[0041]
[0042] This exa...
Embodiment 2
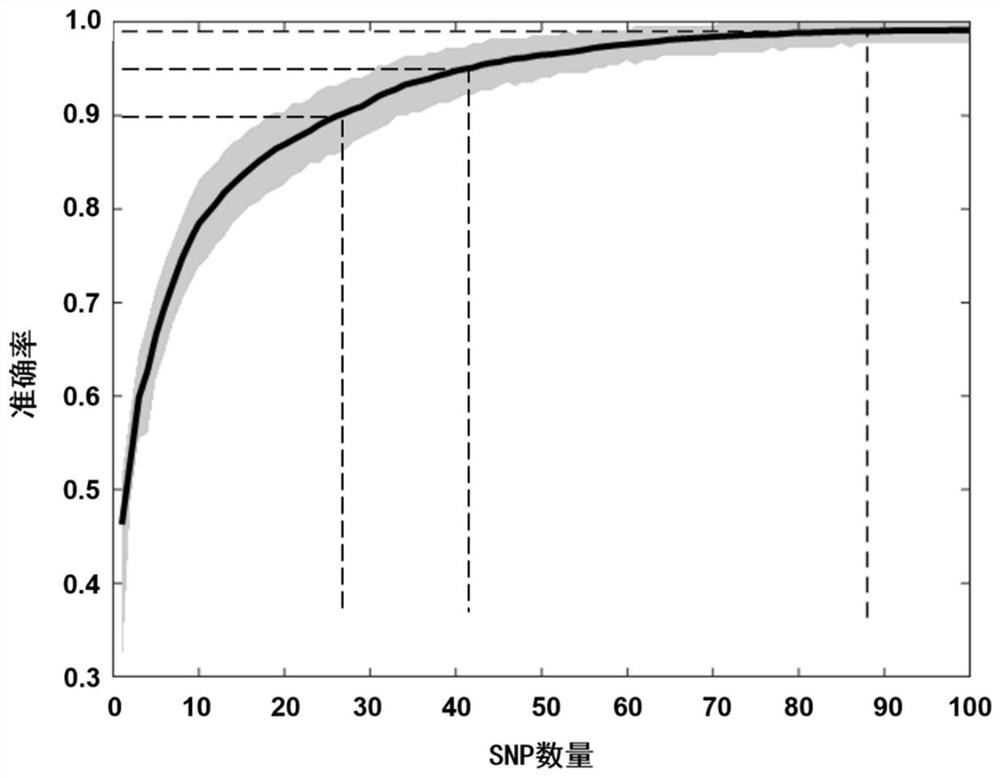
[0051] Example 1 extracts SNP combinations that are beneficial for ancestry inference from a total of 150,793 SNPs. The algorithm can weigh the ancestry inference ability of the SNP itself and the information overlap between different SNPs to obtain the best combined inference effect. Add the screened SNPs one by one, and calculate the average classification accuracy rate AAC, and get the curve as figure 1 shown. The classification accuracy AC is defined as the ratio of the number of correctly classified samples to the total number of test samples,
[0052]
[0053] The average classification accuracy (AAC) is defined as the average value obtained by repeatedly calculating the AC value 1000 times when the test set is randomly selected.
[0054] In this example, three methods are used to evaluate the performance of the SNP reference system obtained in Example 1. The first way is to directly compare the real ancestry with the predicted ancestry; the second way is to calcul...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com