Protein fusion design method for fusing two proteins based on alpha helix and maintaining activity of respective subunits
A protein and α-helix technology, which is applied in the direction of fusion polypeptide, chemical instruments and methods, and the use of vectors to introduce foreign genetic material, etc., can solve problems such as cumbersome recombinant expression, difficulty in designing mutation sites, and difficulty in protein expansion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Structure and sequence comparison of tubulin from different species
[0028] 1. Through the PDB database, retrieve microtubules from four different sources: Saccharomyces cerevisiae, Eurasian wild boar (Sus scrofa), Homo sapiens and Proteus (Prosthecobacterdejongeii), and collect their structure and sequence information;
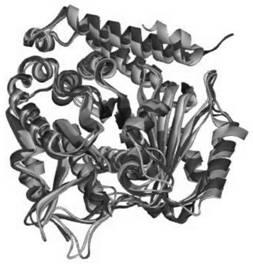
[0029] 2. Compare the amino acid sequence homology of microtubule subunits by DNAMAN software: such as figure 1 As shown, the homology degree of α subunit is 70.65%;
[0030] 3. Compare the structural similarity of microtubule subunits by Discovery Studio2.5 software: such as figure 2 As shown, the α subunits of different species are very similar;
[0031] 4. Considering the convenience of experimental operation, convenience of expression and purification, and protein expression, the microtubule α subunit of Protecobacter dejongeii was selected as the basic protein; the D domain of protein A can bind to the antibody of the VHⅢ family. An...
Embodiment 2
[0032] Same amino acid substitution in embodiment 2α-helical structure
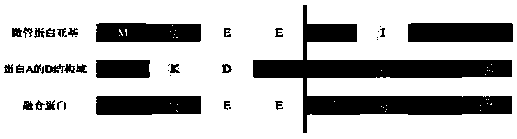
[0033]1. Analyze the structure of the 410-435 amino acid sequence of the C-terminal (remove the C-terminal carboxyl structure) of the tubulin α subunit of Protecobacter dejongeii, which is an α-helical structure; analyze the D domain of protein A Structure, found that it is three consecutive α-helical structures;
[0034] 2. Using the α-helical structure for splicing, the C-terminus of the tubulin α-subunit of Prosthecobacterdejongeii is connected to the N-terminus of the D domain of protein A by α-helical connection, and the same amino acid substitution is used. Connection, same amino acid substitution see image 3 hint.
Embodiment 3
[0035] α-helix formation condition screening of embodiment 3 fusion mode
[0036] 1. The conformation of the main chain of the peptide chain can usually be described by Ω, Φ and Ψ, the angle generated by the rotation around the C-N bond in the amide bond is called Ω, and the dihedral angle around the Cα-N bond axis (C-N-Cα-C) is called Φ, and the dihedral angle (N-Cα-C-N) around the Cα-C bond axis is called Ψ.
[0037] Because the C-N bond in the amide bond is affected by the C=O bond, it has a partial double bond property, and because the trans conformation has lower energy, Ω is usually about 180°. The α helix consists of 3.6 residues per turn, the pitch is 0.54nm, Φ is about -57°, and Ψ is about -47°;
[0038] 2. Use the Chimera software to conduct preliminary screening of the same amino acid site splicing at the C-terminal of the tubulin α subunit and the N-terminal of the D domain of protein A, and combine the Ω, Φ and Ψ numerical analysis to screen out a more reasonable...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap