Detecting breast cancer
A technology for cancer and biological samples, applied in the field of breast cancer detection, which can solve problems such as inability to accurately identify women
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment I
[0444] This example describes the discovery and tissue validation of breast cancer specific markers.
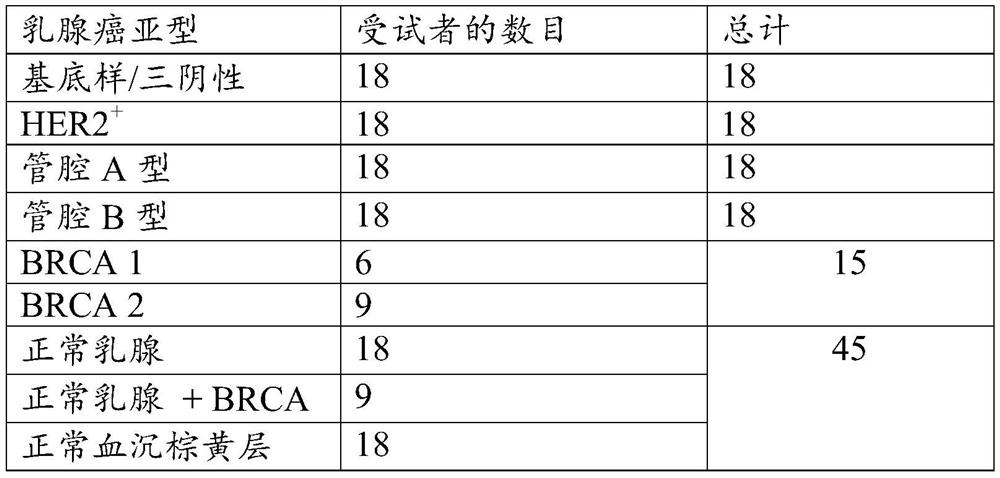
[0445] Table 1 shows the number of tissue samples for each breast cancer subtype used in the discovery of breast cancer specific markers.
[0446] Table 1.
[0447]
[0448] For the discovery of methylation markers by RRBS, frozen tissue samples were obtained from 72 invasive breast cancer cases (18 luminal A, 18 luminal B, 18 basal-like / triple negative and 18 HER2+), 15 Aggressive breast cancer from patients with germline BRCA mutations (6 BRCA1, 9 BRCA2) and 45 controls (18 normal breasts (reducing mammoplasty or preventive mastectomy), germline BRCA carriers in Nine histologically normal breasts (prophylactic mastectomy) and 18 normal buffy coats were obtained. Tumor and breast tissue sections were reviewed by an experienced gastrointestinal pathologist to confirm the diagnosis and estimate abnormal cellularity. The sections were then subjected to macroscopic dissect...
Embodiment II
[0528] This example describes tissue validation of breast cancer specific markers.
[0529] Individual tissue samples (fresh frozen) were selected from the Institutional Cancer Registry at the Mayo Clinic Rochester and reviewed by experienced pathologists to confirm correct classification and to guide macroscopic dissection. Cases included 29 triple-negative / basal-like invasive breast cancers, 34 HER2 invasive breast cancers, 36 luminal A invasive breast cancers, and 25 luminal B invasive breast cancers. Also included were 5 BRCA1 cancers and 6 BRCA2 cancers, 21 HGD DCIS and 12 LGD DCIS. Controls included 27 age-matched normal breast tissue and 18 buffy coat samples from normal women.
[0530] The 55 methylated DNA markers (MDMs) were selected from a list of 80 MDMs tested with discovery samples (see Example 1 and Tables 11-15).
[0531] Genomic DNA was prepared using the QIAamp DNA Mini Kit (Qiagen, Valencia CA) and bisulfite transformed using the EZ-96 DNA Methylation Kit ...
Embodiment III
[0557] This example describes the identification of breast tissue and plasma markers for the detection of breast cancer.
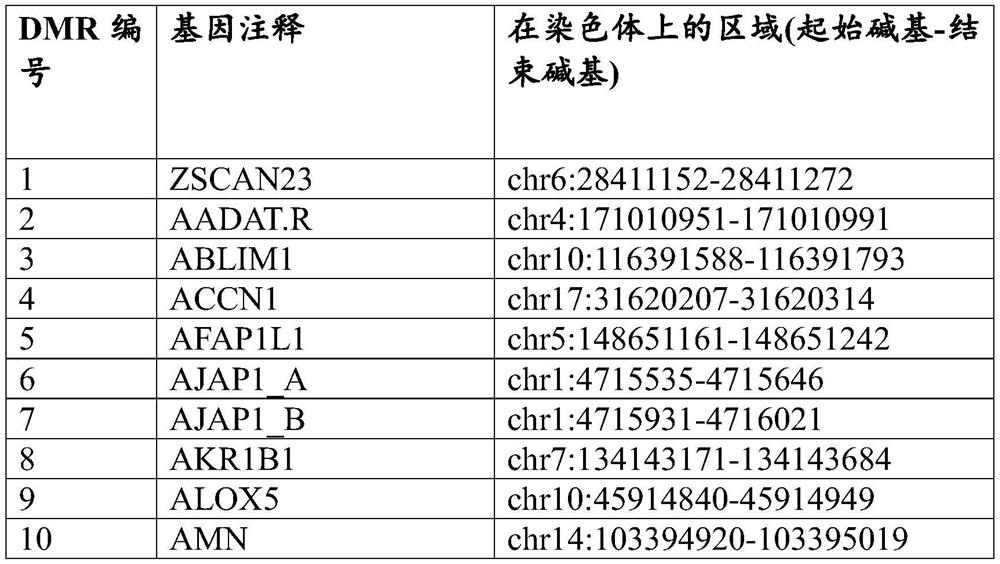
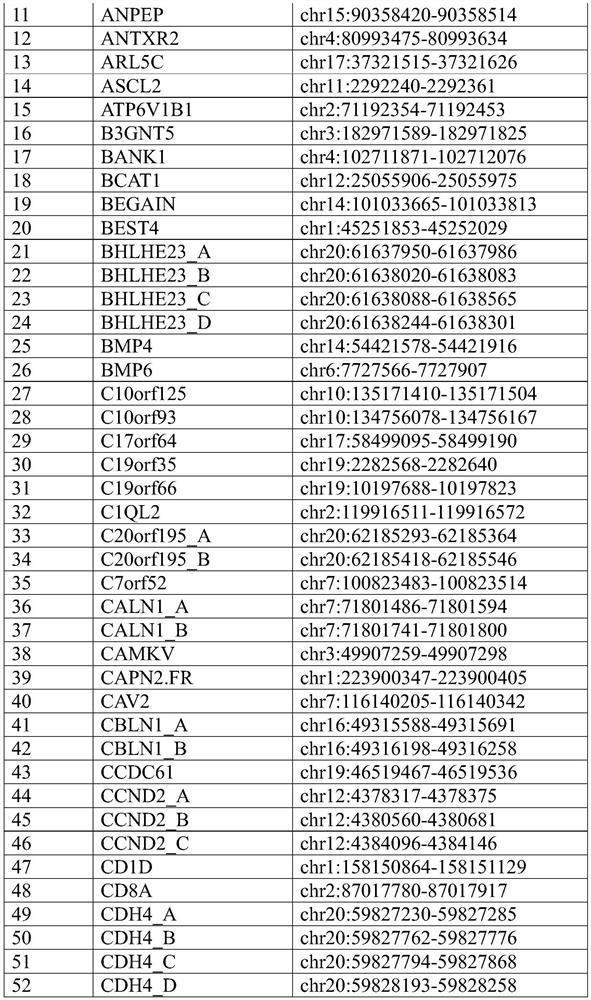
[0558] Candidate methylation markers for detection of breast cancer were identified by RRBS on breast cancer and normal breast tissue samples. 58 markers were initially identified, and target-enriched long probe quantitative amplification signal assays were designed and ordered (for general techniques, see e.g. WO2017 / 075061 and US Patent Application Serial No. 15,841,006) (Table 18 shows the Methylated regions with normal breast tissue) (Table 19 and Table 20 show the primer and probe sequences for the markers shown in Table 18). After design screening and redesign, 56 markers (see Table 21) were selected and assayed, triplicated and tested with tissues. Assays were split equally between FAM and HEX reports and triplexed with reference assay B3GALT6 reporting on Quasar670.
[0559] Table 18. Methylated regions that distinguish breast cancer tissue from ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com