Diagnostic biomarkers and therapeutic targets for pancreatic cancer
a pancreatic cancer and biomarker technology, applied in the field of cancer diagnostics, prognostics, and therapeutics, can solve the problems of low survival statistics, limited finding of t cell antigens, ineffective treatment, etc., and achieve the effect of long-term disease-free survival
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Materials and Methods
Patients, Serum and Tissue Samples
[0044]Patients were enrolled in a phase II study of an allogeneic GM-CSF secreting whole cell pancreatic cancer vaccine in compliance with the Johns Hopkins Medical Institution Institutional Review Board (IRB)-approved J9988 protocol. Blood samples were collected pre-vaccination, 14 days after 1St vaccination and 28 days after each subsequent vaccination. Sera was collected by centrifugation, aliquoted and stored at −80° C. Pancreatic tumor tissue samples were obtained from patients prior to vaccination.
Antibody Purification
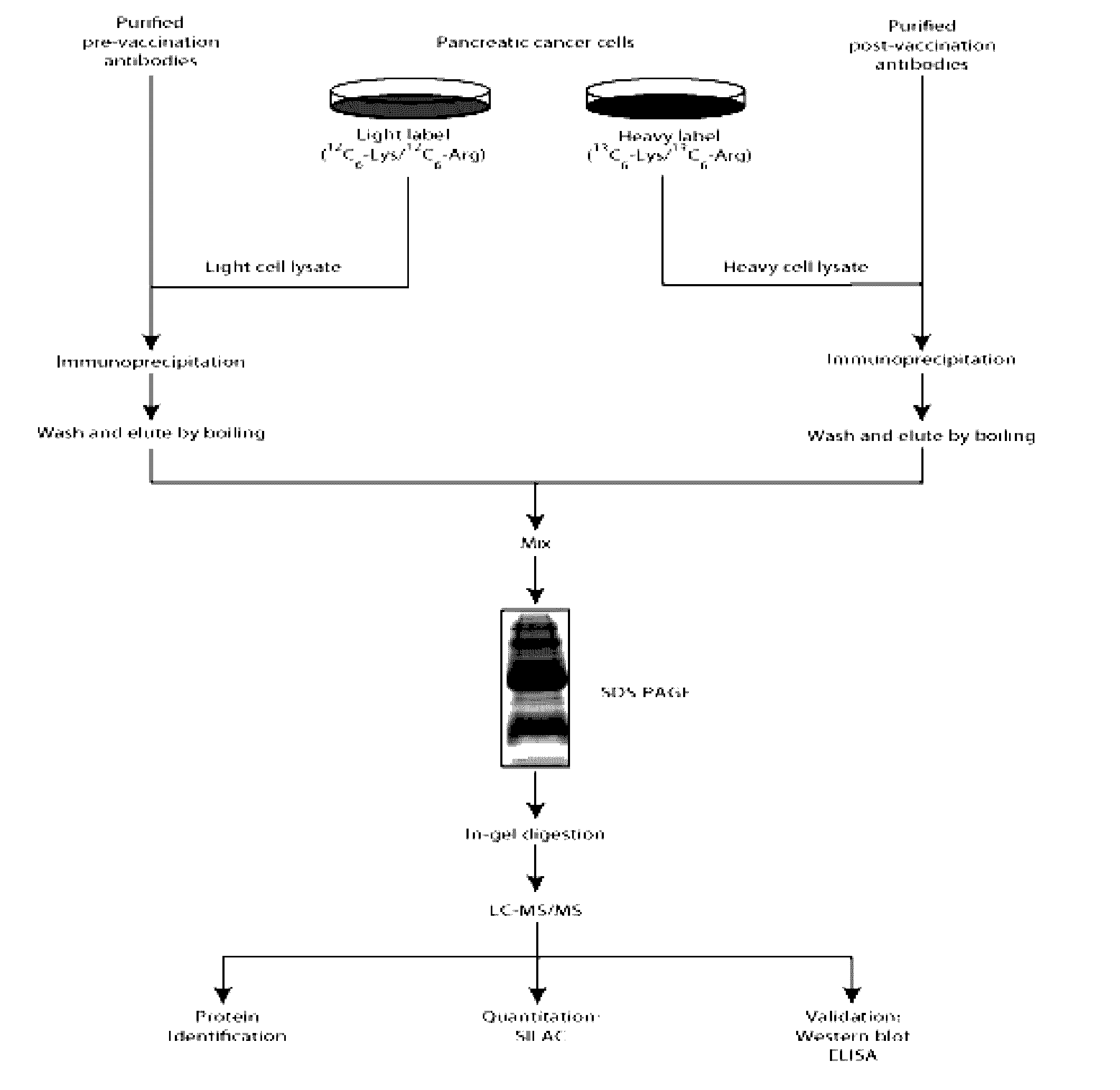
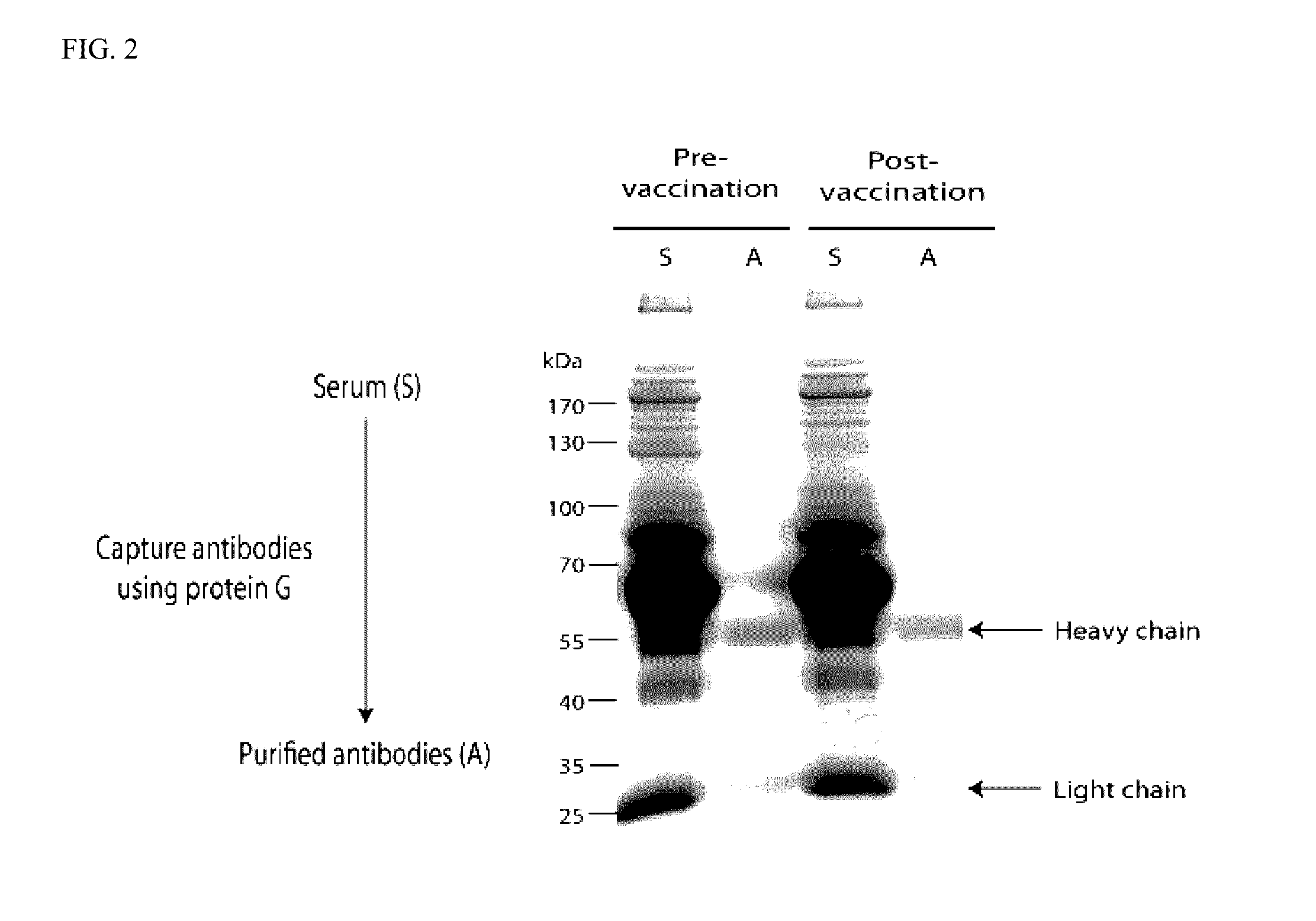
[0045]Antibodies were purified from pre- and post-3rd vaccination sera using a protein G column (GE Healthcare, Piscataway, N.J., USA) as per manufacturer's protocol. Quantification of purified antibodies was done using NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, Mass., USA).
Sample Preparation
[0046]The human pancreatic cancer cell line, Panc 10.05 was grown as previously described. For the ...
example 2
Design and Validation of Quantitative Proteomic Approach
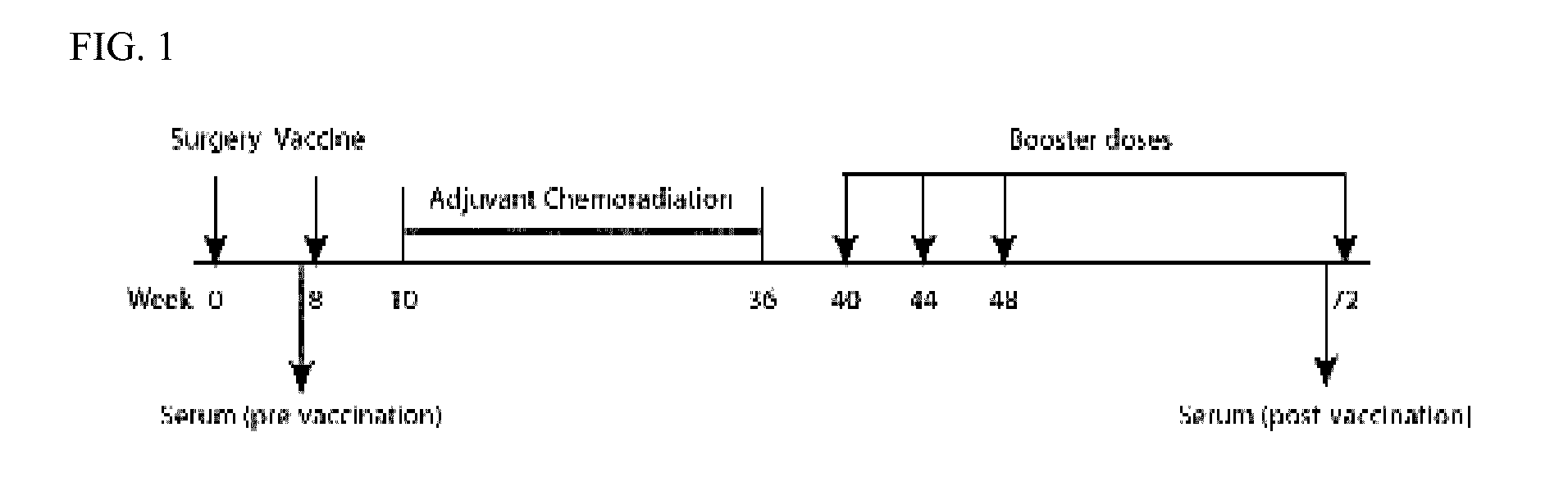
[0052]60 pancreatic cancer patients, who had their pancreas surgically removed, were involved in the study (FIG. 1) (2). The patients received their first vaccination 2 months after surgery. One month after the first vaccination, the patients underwent a 6-month course of chemoradiation. The second, third and fourth vaccines were each administered at sequential one-month intervals from the time of chemotherapy completion. The fifth, and final, vaccination was received 6 months after the fourth vaccination. Serum samples were obtained pre- and post-vaccination for all five vaccinations (2). The 60 vaccinated patients were divided into 3 groups (A, B and C) based on length of disease free survival (DFS) (2). Group A was composed of 12 patients who received all of the scheduled vaccinations and demonstrated a DFS>3 years (prolonged DFS as well as overall survival). The clinical time point cutoff was determined to be 3 years becaus...
example 3
Identification of Proteins by the SASI Approach
[0053]To identify the proteins in the post-vaccination sera of patients in Group A (DFS>3 years), we used the immunized sera from three patients (patients 9, 27 and 52) who demonstrated other evidence of post-vaccination immune responses. We identified a total of 976 proteins for patient 9, 811 proteins for patient 27 and 727 proteins for patient 52 (FIG. 5). A broad range of post-vaccination antibody response was observed; from a 16 fold change increase post-vaccination to a 10 fold change decrease. The majority of the proteins, as expected, had no change in response post-vaccination. We identified 51 proteins for patient 9, 47 proteins for patient 27 and 54 proteins for patient 52 that had a 2 fold change in response. Through the SASI approach, we present the first large scale study to identify and categorize proteins that are targeted by antibodies in the human body.
[0054]Pre-vaccination and post-4th vaccination sera from 3 patients,...
PUM
| Property | Measurement | Unit |
|---|---|---|
| mass tolerance | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| thick | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com