Method for the analysis of breast cancer disorders
A breast cancer, disorder technology with applications in biology and chemistry, molecular biology and human genetics to address problems such as poor performance characteristics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
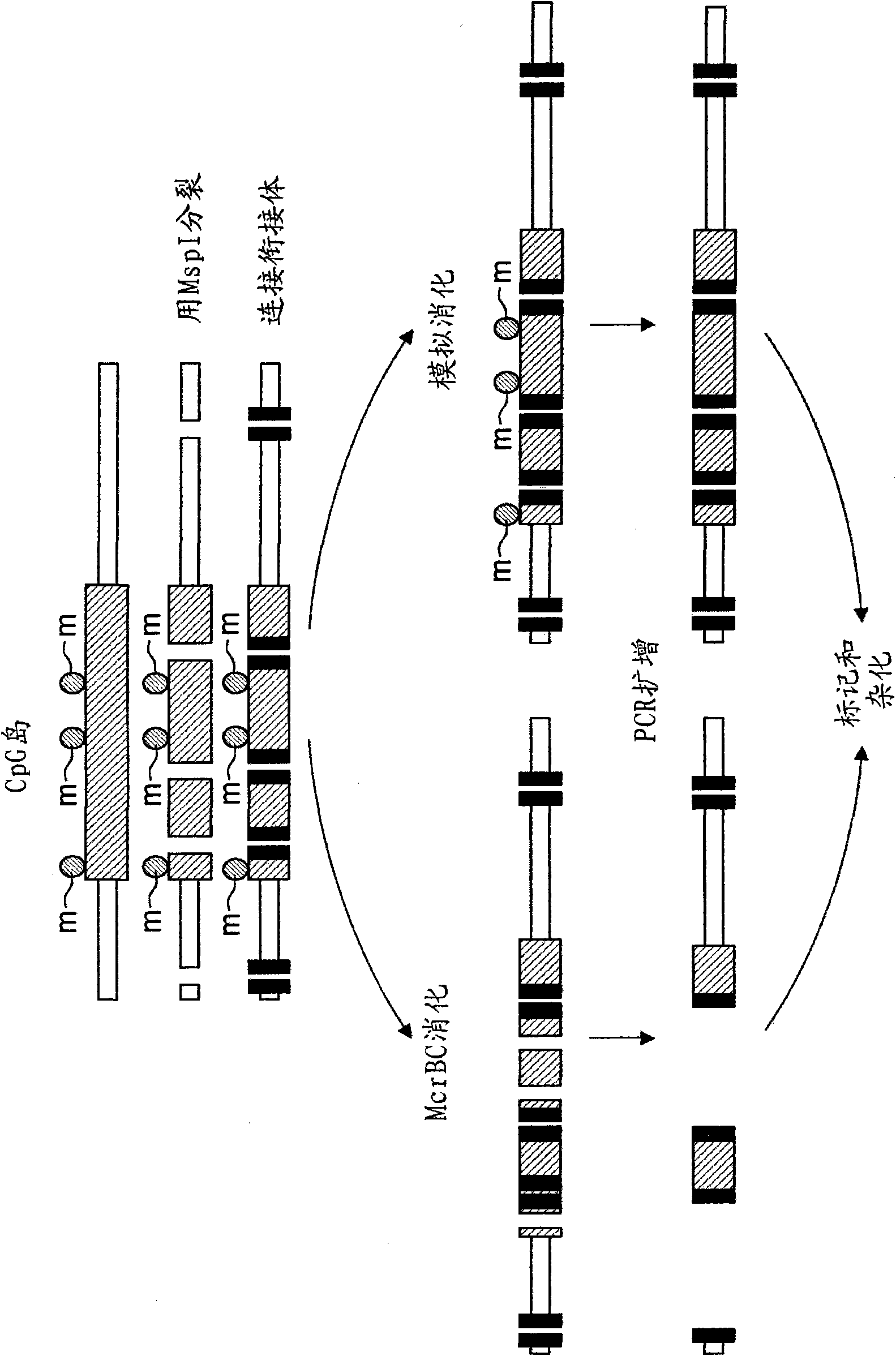
[0018] The inventor surprisingly found that a small part of the DNA sequence can be used to analyze breast cancer disorders. This is accomplished by determining the genome methylation status of one or more CpG dinucleotides in the sequence disclosed herein or its reverse complement. A total of 900 sequences suitable for this analysis were identified. It turns out that 100 sequences are particularly suitable.
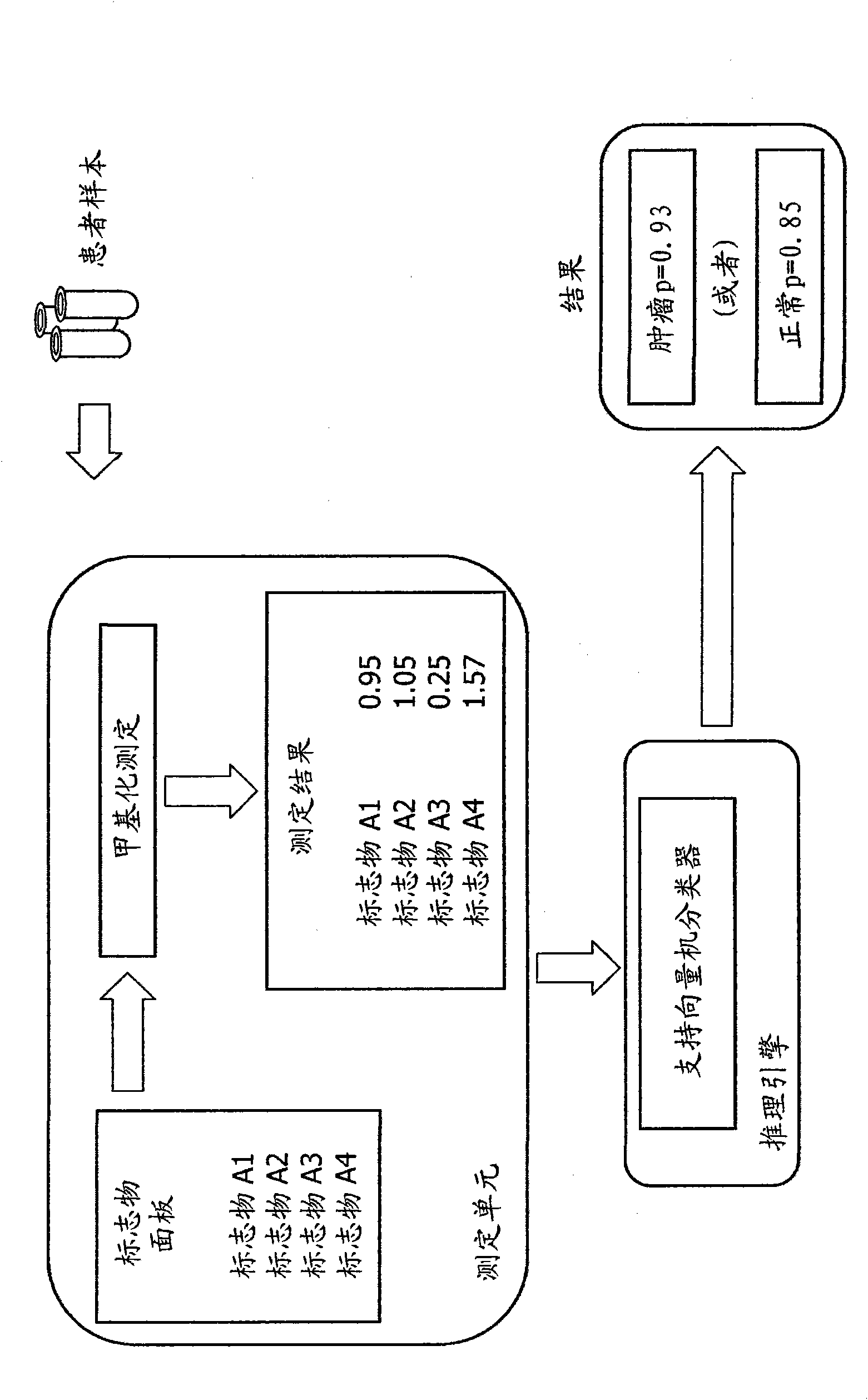
[0019] Based on only 10 sequences, such as the first ten features from Table 1A or 1B (p value 0.000.1), a classification accuracy of 94% can be achieved (Comparison of the total number of correct predictions on whether a given sample is from a breast tumor Total number of predictions made, 49 / 52). The sensitivity for tumor detection is 92.5% (37 / 40), and the specificity for tumor detection (specificity = 100%). Increasing the feature size to 50 gives a classification rate of 96% (50 / 52 is accurately classified).
[0020] The sequence can be found in the gene as can be se...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com