Clear cell renal cell carcinoma biomarkers
A biomarker, clear cell technology, applied in the determination/examination of microorganisms, biochemical equipment and methods, instruments, etc., which can resolve tissue changes, lack of methods to assess the efficiency of systemic treatment in patients with advanced clear cell renal cell carcinoma, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088] Example 1 - Materials and methods
[0089] patient information
[0090] Fresh-frozen normal tumor tissue was obtained from nephrectomy cases with institutional research ethics review board approval and patient consent. Normal tissue is collected from a site away from the tumor. Table 1 relates to detailed patient information for this study.
[0091] ID age at diagnosis Race gender Tissue type Fuhrman nuclear grade 74575859 80 China female clear cell carcinoma Class II 17621953 48 China male clear cell carcinoma Class II 57398667 56 China male clear cell carcinoma Class III 70528835 52 China male clear cell carcinoma Class III 77972083 50 China male clear cell carcinoma Class IV 75416923 49 unknown male clear cell carcinoma Class II 20431713 65 India male Clear cell carcinoma with papillary features Class IV 40911432 56 China female clear cell carcin...
Embodiment 2
[0215] Example 2 - Abnormal cis-regulatory patterns in clear cell renal cell carcinoma tumors
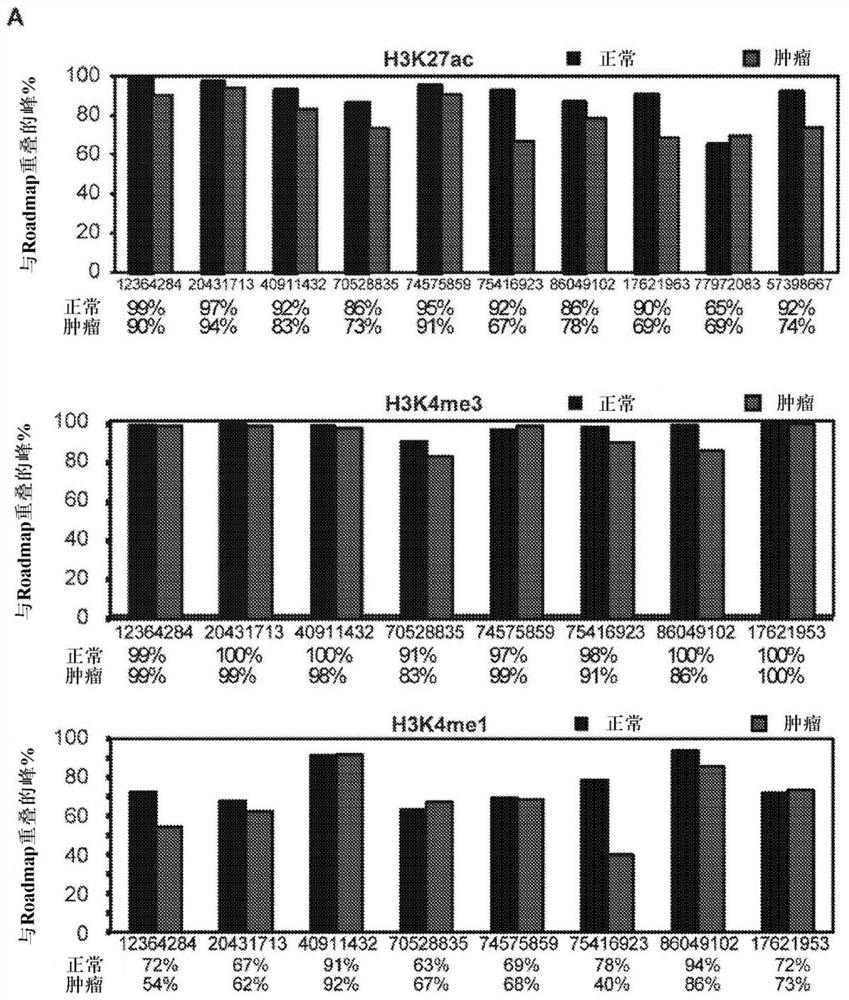
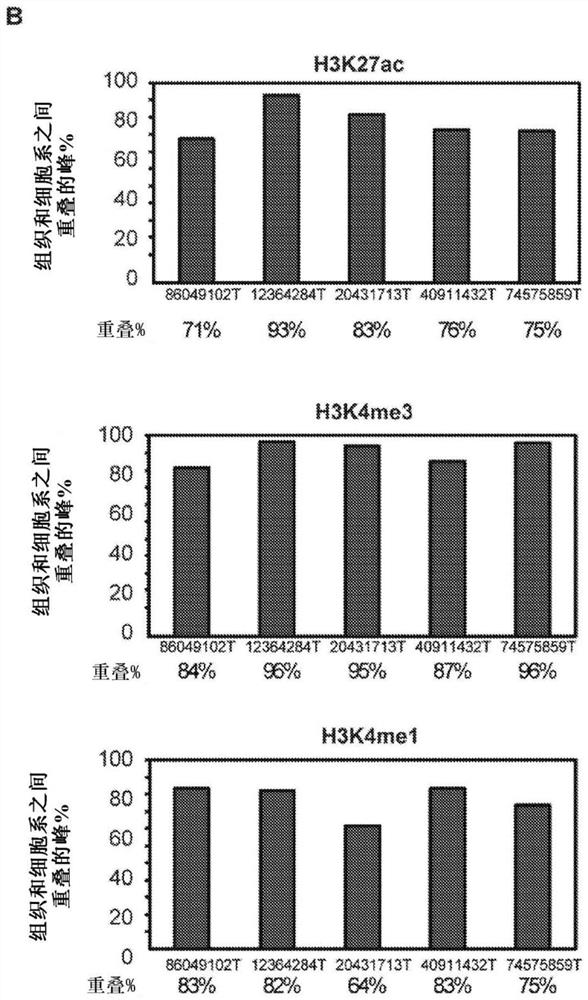
[0216] To explore whether clear cell renal cell carcinoma tumors exhibit changes in their cis-regulatory patterns in vivo, 10 primary tumor / normal pairs, 5 patient-matched tumor-derived cell lines, 2 commercially available Histone chromatin immunoprecipitation sequencing (ChIP-seq) profiles (3 markers) were generated in clear cell renal cell carcinoma lines (786-O and A-498) and 2 normal renal cell lines (HK2 and : H3K27ac, H3K4me3 and H3K4me1). Table 1 in Example 1 shows the clinical information of the patients. Of the original 87 samples, 79 samples passed the pre-sequencing quality control filter and were subjected to ChIP-seq processing and downstream analysis. A total of 2,363,904,778 uniquely mapped reads were generated. On average 89% of H3K27ac peaks, 98% of H3K4me3 peaks and 76% of H3K4mel peaks obtained in our normal kidney tissue overlapped with peaks from adult kidney...
Embodiment 3
[0222] Example 3 - Tumor-specific enhancers are associated with hallmarks of clear cell renal cell carcinoma
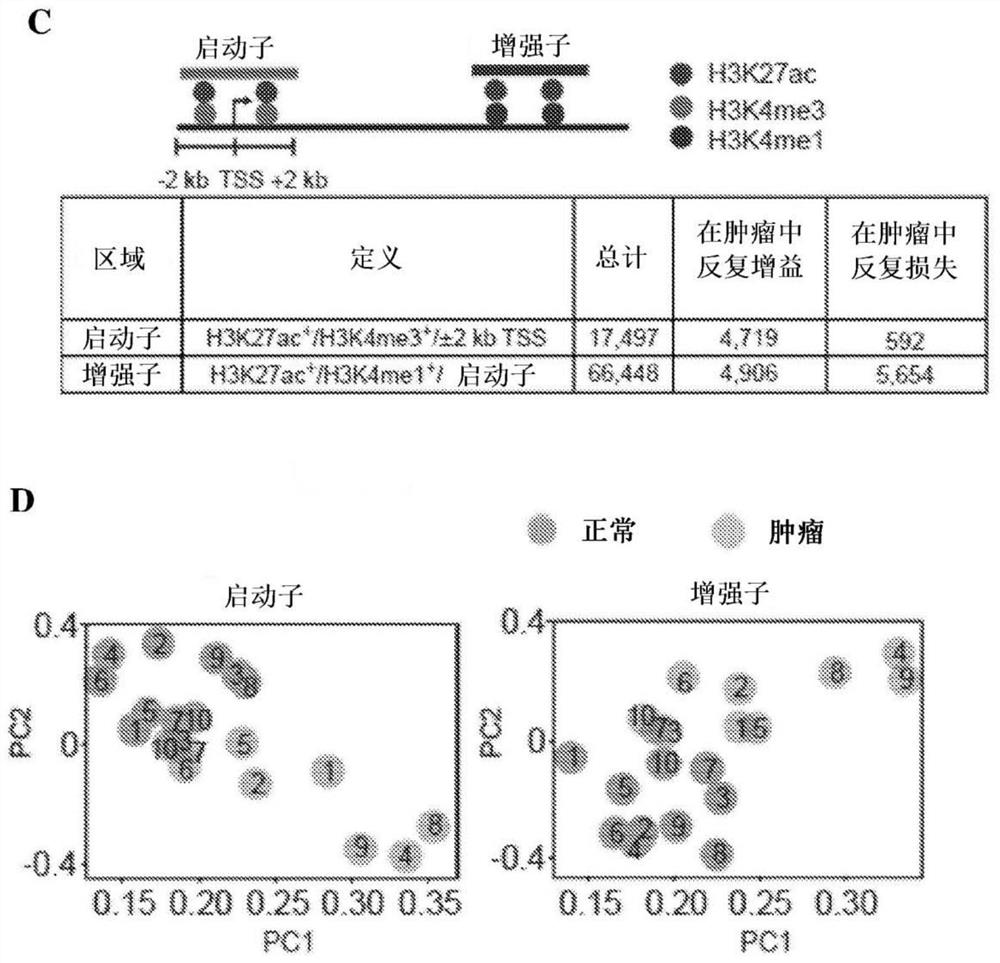
[0223] To identify genes regulated by tumor-specific regulatory elements, three methods were used to assign enhancers. The first approach utilizes a predefined linear proximity rule (GREAT algorithm) involving a set of high-confidence genes. MSigDB pathway analysis using GREAT-assigned genes revealed that the gain-in enhancer exhibited a highly significant renal cell carcinoma-specific signature compared to the gain-in promoter (enhancer q-value = 3.2 x 10 -26 ; promoter q value = 1.5 x 10 -1 , binomial FDR; figure 2 A). While the gain promoters are involved in general cancer processes (e.g. cell cycle, transcription, and RNA metabolism against a complete list of promoter pathways), the gain enhancers are enriched for disease-specific features of clear cell renal cell carcinoma, including targeting Complete list of enhancer pathways for HIF1α network activity, pr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com