Epigenetic regulatory complex for control of gene expression
A technology for polypeptide complexes and expression vectors, which is applied in the field of compositions with histone methyltransferase activity and can solve problems such as failure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0075] Example 1: Identification of Blimp1 activity
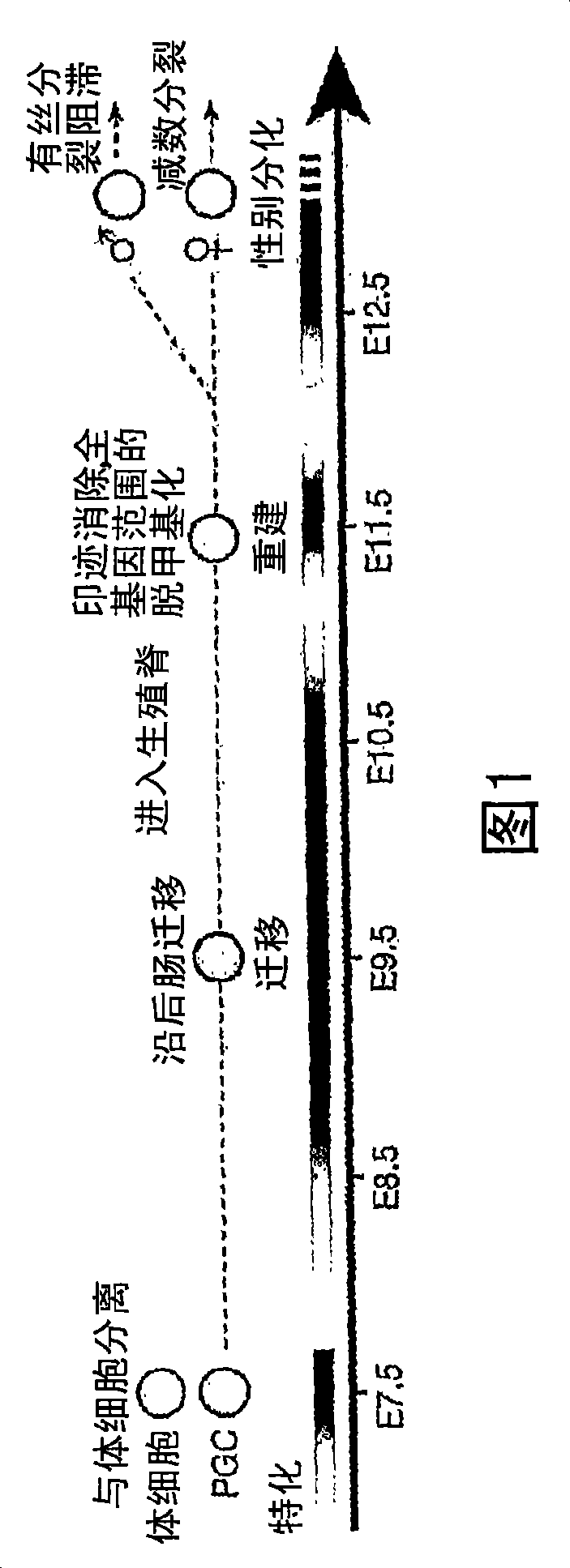
[0076] Significant epigenetic modifications occur immediately after mouse PGC specification, including methylation and acetylation of histone termini by methyltransferases (HMTases) and acetyltransferases (HATs), respectively. Among the candidate genes that may regulate these epigenetic changes in PGCs are HMTases belonging to the conserved SET / PR domain protein family. Expression analysis of 25 candidate genes containing SET / PR domains was performed in E7.5 PGCs and peripheral somatic cells, and Blimp1, G9a, Set1, Ezh2, and Pfml were found to be expressed in embryonic regions containing PGCs ( figure 1 b and Figure 7 ). However, only Blimp1 expression was restricted to PGCs at E7.5 and persisted in germ cells thereafter.
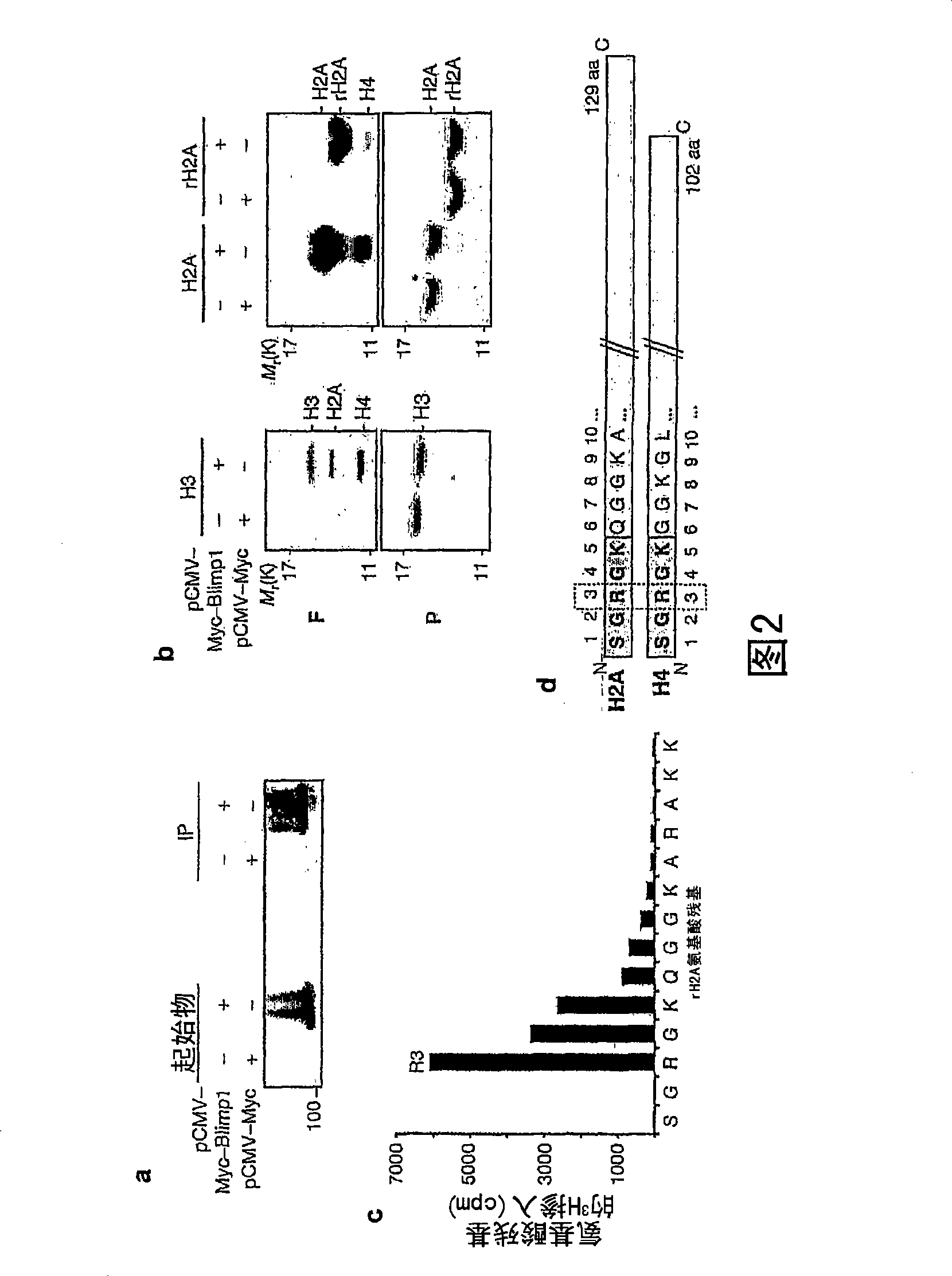
[0077] To investigate the activity of the Blimpl SET / PR domain, it was first determined that the transiently expressed Myc-tagged mouse Blimp1 in 293T cells could be efficiently immunoprecipitated us...
Embodiment 2
[0079] Example 2: Identification of the Blimp1 / Prmt5 complex
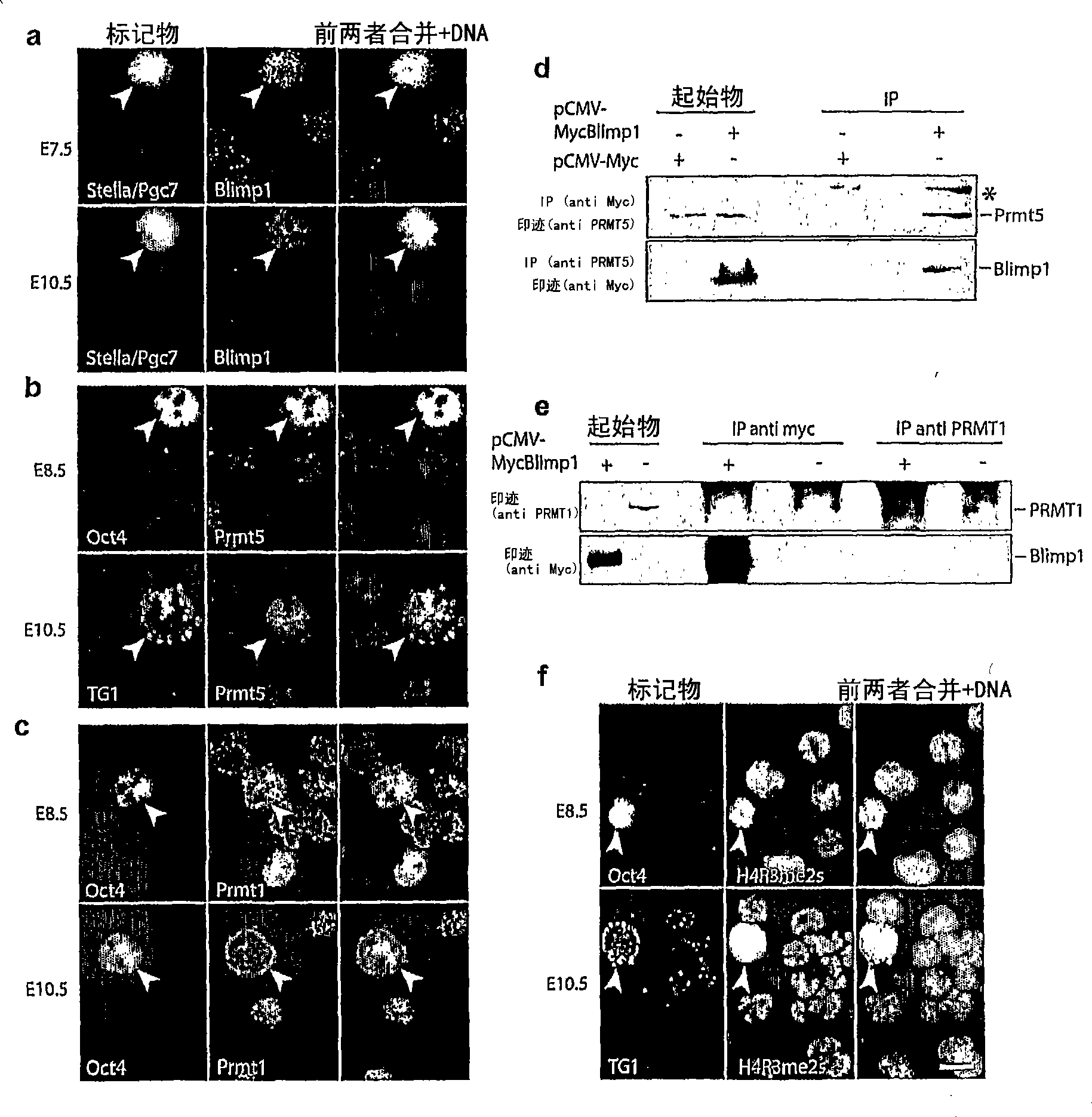
[0080] Since the SET / PR domains are associated with histone methyltransferase activity only on lysine residues, it can be assumed that the arginine methyltransferase activity detected above may not be attributable to Blimp1 and must imply Other HMTases exist. It has been previously reported that two protein arginine methyltransferases, Prmt1 and Prmt5, mediate the methylation of histone H4 R3. Prmt1 is a type I arginine methyltransferase that produces NG-monomethylarginine (Rme1) and asymmetric NG,N'G-dimethylarginine (Rme2a) on different types of substrates. As mentioned above, Prmt5 belongs to the type II arginine methyltransferases responsible for monomethylation of arginine (Rme1) and symmetric NG, N′G-dimethylation (Rme2s) ( Figure 8 ) 20 . Expression of these proteins in nascent PGCs and peripheral somatic cells was analyzed using the single-cell cDNA library described above to test whether one of these ...
Embodiment 3
[0082] Example 3: Activity of the Blimp1 / Prmt5 complex in vivo
[0083] The specificity of the antibody raised against H4R3me2s was tested, and the results showed that the antibody could efficiently recognize histones H2A and H4 from calf thymus ( Figure 8 b Left; H2A and H4 also appear as contaminants in H3 and H2B preparations). In a competition assay, this antibody was efficiently titrated by the R3me2s-containing H4(1-9C) polypeptide (Fig. S2c). In addition, this antibody also recognized the Blimp1 / Prmt5 HMTase assay product of H4 and H2A (Fig. S2b right), thus further demonstrating that the complex is responsible for the symmetric dimethylation activity of Prmt5 rather than the asymmetric dimethylation activity of Prmt1. methylation activity.
[0084] PGCs isolated from early embryos were analyzed by immunocytochemistry using an antibody specific for H2 / H4R3me2s modification. Significant H2A / H4R3me2s were seen in both PGCs and somatic cells at E8.5 ( image 3 f), but...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com