Method for synthesis of double-stranded DNA corresponding to RNA, and method for amplification of the DNA
A synthesis method and corresponding technology are applied in the field of double-stranded DNA synthesis, which can solve the problems of high-priced instruments, inability to analyze, and inability to obtain cDNA, and achieve the effects of simple operation and low price.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
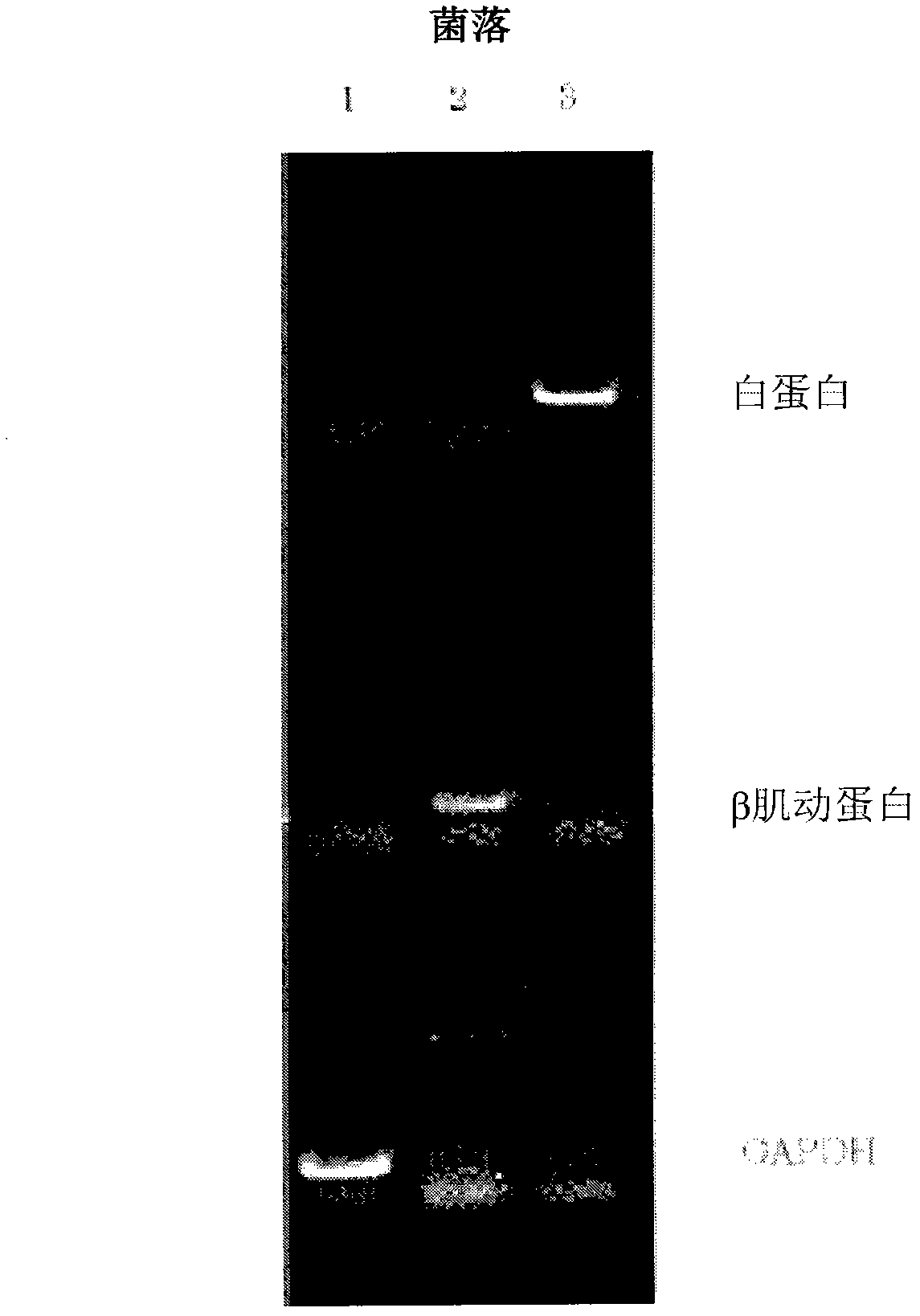
[0053] Example 1 Cloning of 3 mRNAs (human albumin, β-actin, GAPDH)
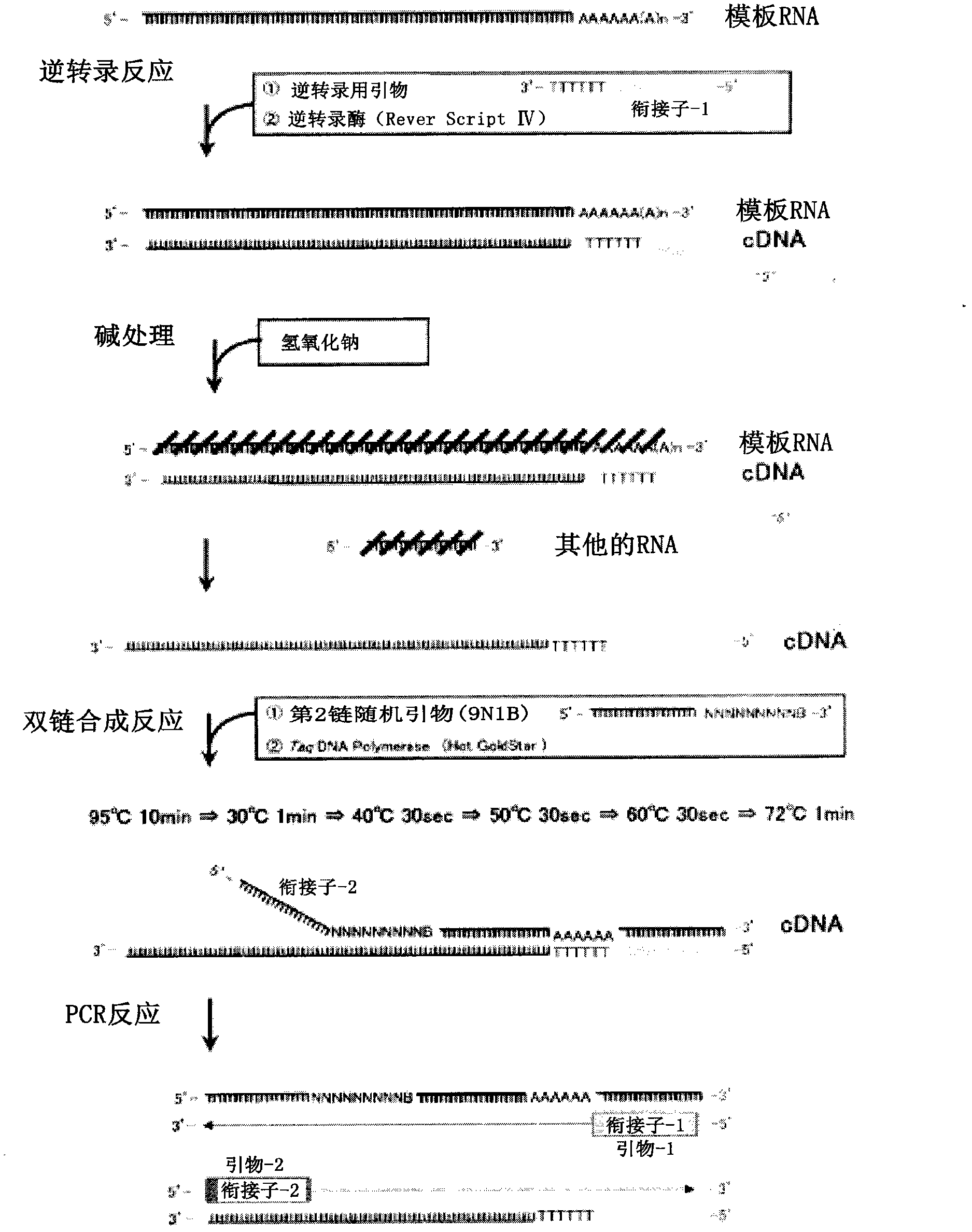
[0054] 1. Reverse transcription reaction
[0055] will respectively contain 1×10 9 Copied human albumin (2122 bases), β-actin (1874 bases), and GAPDH (1380 bases) mRNA (manufactured by NIPPON GENE CO., LTD.) aqueous solution 11 μL , as template RNA in water. To this aqueous solution was added 1 µL of a 20 µmol / L primer solution for reverse transcription (5'-GACCATATGACGAGATCCGAGCTTCTTTTTTTTTTTTTTTTTTTTTT-3'). Next, it was heated at 70° C. for 3 minutes to thermally denature it, and then left to stand in an ice bath for 2 minutes. Then, add: 2 μL reverse transcription reaction buffer (containing 750 mmol / L potassium chloride, 30 mmol / L magnesium chloride, 500 mmol / L Tris hydrochloric acid buffer solution (pH 8.3) of 50 mmol / L dithiothreitol), 4 μL dNTPs ( Respectively 2.5mmol / L of dATP, dGTP, dCTP, dTTP solution mixed solution, produced by Nippon Gene Company), 1 μL ribonuclease inhibitor (20 units / μL, ...
Embodiment 2
[0164] Example 2 Different amplification effects brought about by different reaction temperatures during the synthesis of the second strand.
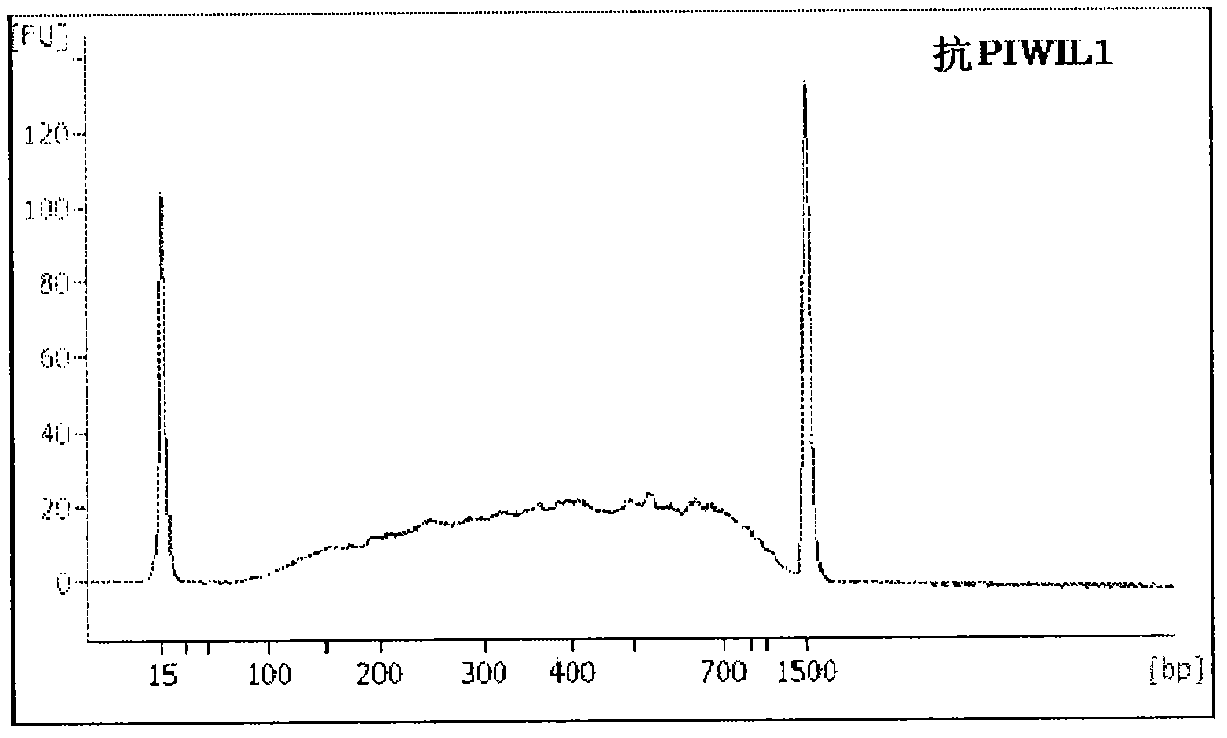
[0165] 1. Acquisition of Ago2-bound RNA from HeLa cells
[0166] (1) Preparation of human anti-Ago2 antibody immobilized carrier
[0167] 1 ml of PBS (pH 7.4) was added to 20 μL of magnetic carrier Dynabeads Protein G (manufactured by InvitroGen), and after mixing, the carrier was extracted using a magnetic stand. Then, a mixture of 5 μL of 1 mg / mL human anti-Ago2 antibody (manufactured by Wako Pure Chemical Industries, Ltd.) and 95 μL of phosphate-buffered saline (PBS pH 7.4) was added to the carrier, and shaken at room temperature for 1 hour. Next, the carrier was extracted using a magnetic stand, washed three times with 1ml PBS (pH 7.4), and finally suspended in 1ml PBS (pH 7.4), and the resulting solution was the human anti-Ago2 antibody immobilized carrier solution.
[0168] (2) Preparation of HeLa cell extract
[0169] to 1×1...
Embodiment 3
[0190] Example 3 is corresponding to the RNA (derived from the Alu RNA of HeLa cells) that does not have the Cap structure DNA synthesis
[0191] 1. Conversion
[0192] To 2 μL of the PCR reaction solution prepared in 5. of Example 2, 1 μL of 20 ng / μL of pGEMTeasy Vector (manufactured by Promega) and 3 μL of DNA Ligation Kit Mighty Mix (manufactured by TAKARA BIO Co., Ltd.) were added, and reacted at 16°C For 30 minutes, insert the cloned cDNA into the vector. Then, using ECOS TM Competent E.coli DH5α (manufactured by Nippon Genetics Co., Ltd.) was transformed into competent cells using the heat shock method at 42°C for 45 seconds. Next, culture was carried out at 37°C for 16 hours in a Luria-Bertani's broth (LB) agar medium containing 100 µg / mL ampicillin sodium.
[0193] 2. Colony PCR
[0194] Add a part of each of the 96 single bacterial colonies on the culture medium to the reaction solution [full volume 10 μL: 1 μL 10× Universal Buffer (manufactured by Japan Gene ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com