A method for expressing restriction endonucleases efficiently through the protection of methylases
A technology of restriction endonuclease and methylase, applied in biochemical equipment and methods, recombinant DNA technology, botanical equipment and methods, etc., can solve the problem of low restriction endonuclease yield, cumbersome purification procedures, The narrow application scope and other problems can avoid the methylase gene screening, simplify the recombinant expression process, and achieve the effect of wide application.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1, a method for efficiently recombinantly expressing a restriction enzyme using the protection of methylase, the methylase is M.Sss I, and the restriction enzyme D is selected from: AatII , AciI, AclI, AfeI, AgeI, AscI, AsiSI, AvaI, BceAI, BmgBI, BsaAI, BsaHI, BsiEI, BsiWI, BsmBI, BspDI, BspEI, BsrFI, BssHII, BstBI, BstUI, BtgZI, ClaI, EagI, FauI, FseI , FspI, HaeII, HgaI, HhaI, HinP1I, HpaII, Hpy99I, HpyCH4IV, KasI, MluI, NaeI, NarI, NgoMIV, NotI, NruI, Nt.BsmAI, Nt.CviPII, PaeR7I, PluTI, PmlI, PvuI, RsrII, SacII , SalI, SfoI, SgrAI, SmaI, SnaBI, TliI, TspMI, XhoI, XmaI, ZraI.
Embodiment 2
[0056] Embodiment 2, the method for utilizing the protection of methylase described in embodiment 1 to express restriction endonuclease efficiently, comprises:
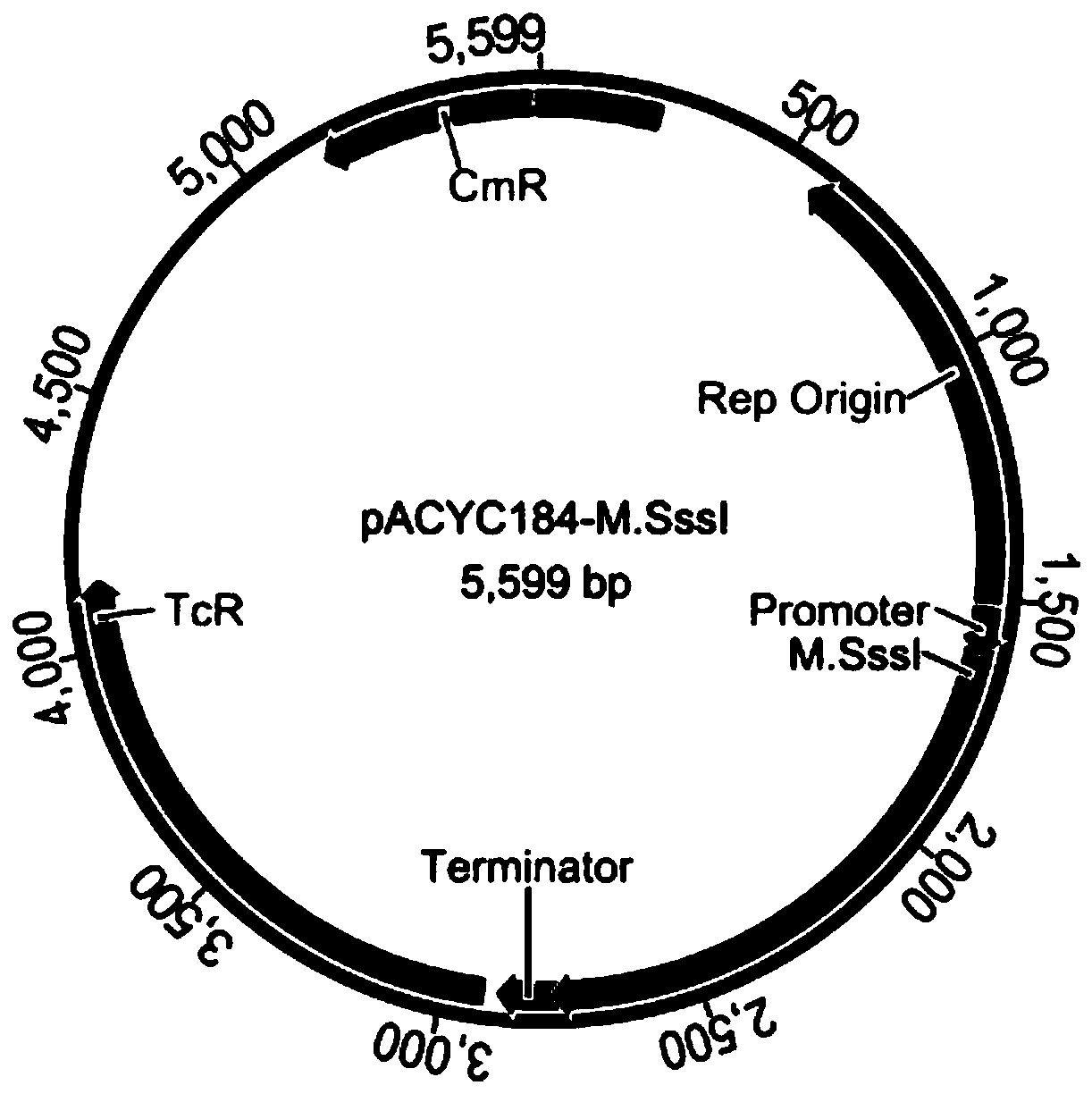
[0057] Step (1): Cloning the M.Sss I gene into the plasmid vector pACYC184 to obtain the M.Sss I constitutive plasmid, transforming the host bacteria, and screening to obtain a positive recombinant strain protected by methylation;
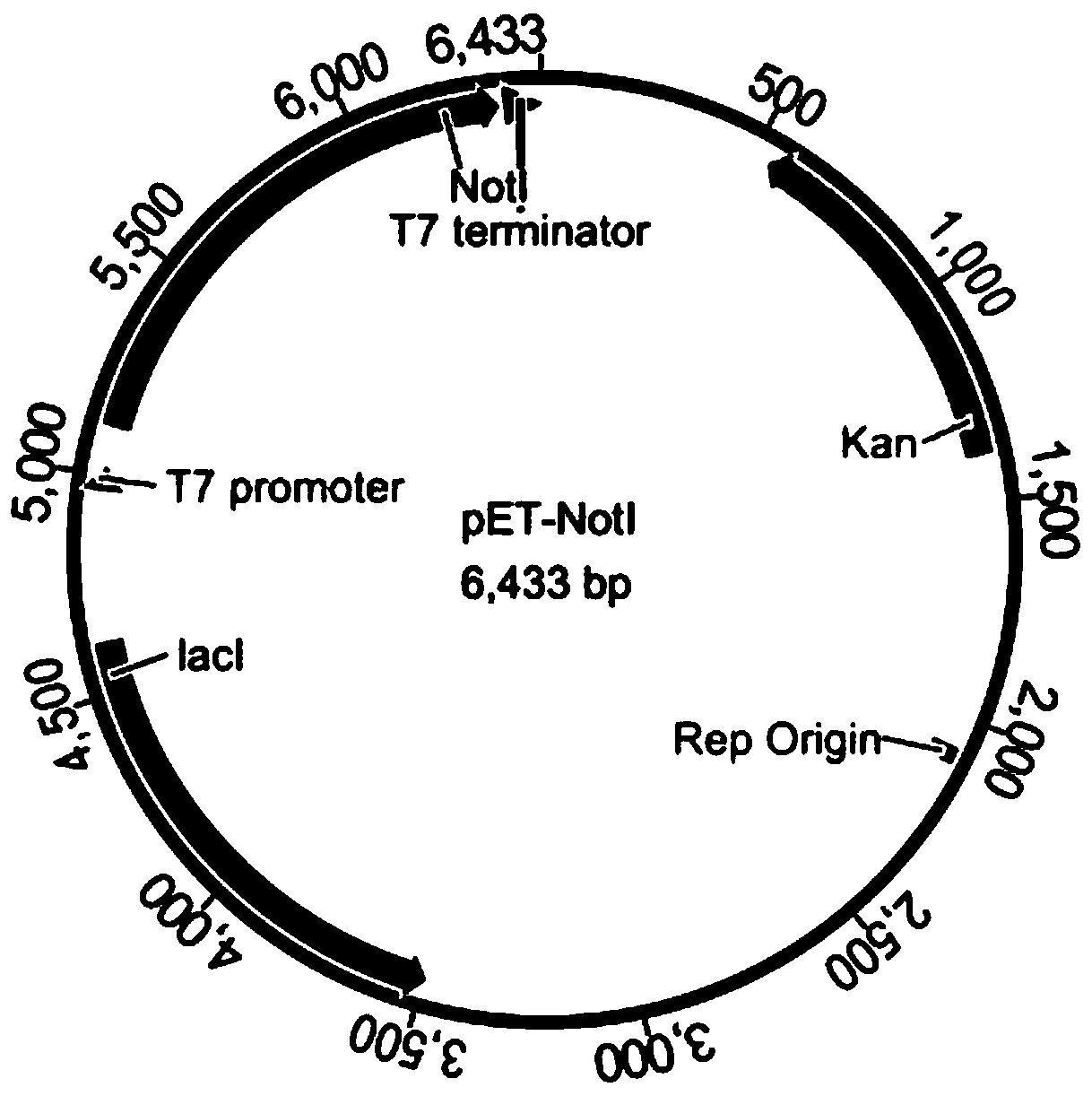
[0058] Step (2): Cloning the restriction endonuclease D gene into the plasmid vector pET-28b to obtain a restriction endonuclease D recombinant plasmid, transforming the methylation-protected positive DNA obtained in step (1) Recombinant bacteria, screening recombinant expression strains of stable, inducible and high-expression restriction endonuclease D.
Embodiment 3
[0059] In Example 3, in the method for expressing restriction endonucleases through high-efficiency recombinant expression using the protection of methylases described in Example 2, the host bacteria are prokaryotic host cells.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com