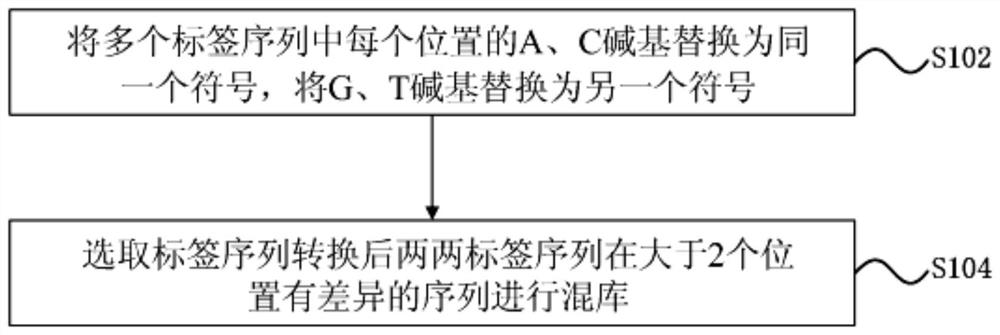
A tag-sequence mixing library method and device for improving the library splitting rate of a sequencing platform
A tag sequence and sequencing platform technology, applied in the field of sequencing, can solve the problem of low split rate of tag sequence sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] The numbers of the two DNA libraries are: WHBRAootMAAFDEAAPEI-30, HUMggzEAAADAAA-129, and the two libraries were mixed on the machine (note: the library name in the example is just a string of symbols used to distinguish different libraries, and has no specific technical meaning; The numbers after the library name, such as 30 and 129 indicate the number of the tag sequence).
[0045] 30. The specific nucleotide sequence of the tag sequence No. 129:
[0046] No. 30: GCTTAATG;
[0047] No. 129: ACAGAGTG.
[0048] Replace the A and C bases with the symbol A, and replace the G and T bases with the symbol B. After the replacement, the information of each tag sequence is as follows:
[0049] No. 30: BABBAABB;
[0050] Number 129: AAABABBB.
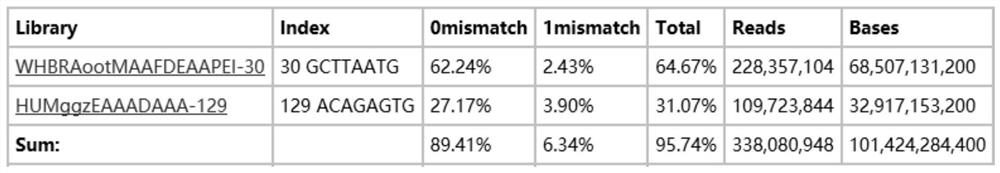
[0051] From the above sequence information, it can be seen that there are 3 different positions between the nucleotides of tag 30 and tag 129 after substitution, image 3 The splitting rate of the actual sequencing tag sequences of t...
Embodiment 2
[0053] For the 4 pepper DNA libraries, the library numbers are: CAPgsdG1AAD96FAAPEI-14, CAPgsdG1ABD96FABPEI-39, CAPgsdG2ADD96FAAPEI-45, CAPgsdG2ACD96FAAPEI-40; according to the data volume requirements, two libraries are required to be mixed on the machine (note: the name of the library in the example) It is just a string of symbols used to distinguish different libraries, without specific technical meaning; the numbers after the library name, such as 14, 39, 45 and 40 represent the number of the tag sequence).
[0054] The specific nucleotide sequences of tag sequences No. 14, 39, 45 and 40 are as follows:
[0055] No. 14: AGAGATCT;
[0056] No. 39: TCCAGTAG;
[0057] No. 45: ACTACAAG;
[0058] No. 40: TTGTCTAG.
[0059] A and C bases are replaced with the symbol A, and G and T bases are replaced with the symbol B. After the replacement, the information of each tag sequence is as follows:
[0060] No. 14: ABABABAB;
[0061] No. 39: BAAABBAB;
[0062] No. 45: AABAAAAB; ...
Embodiment 3
[0072] The numbers of the two DNA libraries are: WHHUMuwoRAAHDEAAPEI-75, WHHUMuwoRAABDEAAPEI-79.
[0073] Specific nucleotide sequences of 75 and 79 tag sequences:
[0074] No. 75: TACTATGA;
[0075] Number 79: CTTATAGA.
[0076] A and C bases are replaced with the symbol A, and G and T bases are replaced with the symbol B. After the replacement, the information of each tag sequence is as follows:
[0077] No. 75: BAABABBA;
[0078] Number 79: ABBABABA.
[0079] From the above sequence information, it can be seen that there are 6 different positions between the nucleotides of tag 75 and tag 79 after replacement, and the two DNA libraries were mixed and sequenced on the machine. Figure 6 The splitting rate of the actual sequencing tag sequences of the two mixed libraries is shown, and it can be seen intuitively that the splitting rate reaches 98.31%.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com