Inhibition of hsd17b13 in treatment of liver disease in patients expressing pnpla3 i148m variation
A technology of I148M and mutation, applied in the field of precision medicine, can solve the problem of not identifying gene variants of chronic liver disease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0332] Example 1: Genetic Interaction Between PNPLA3 rs738409 (p.I148M) and HSD17B13 rs72613567 - Study Design
[0333] In this study, exome sequencing was used to identify Variants associated with serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels as markers of hepatocellular injury. Associations between implicated genetic variants and clinical diagnoses of chronic liver disease were also investigated in DiscovEHR and two independent cohorts. The association between one of these variants and the histopathological severity of liver disease was also investigated in an independent cohort of obese surgical patients undergoing liver biopsy.
[0334] Study Design and Participants
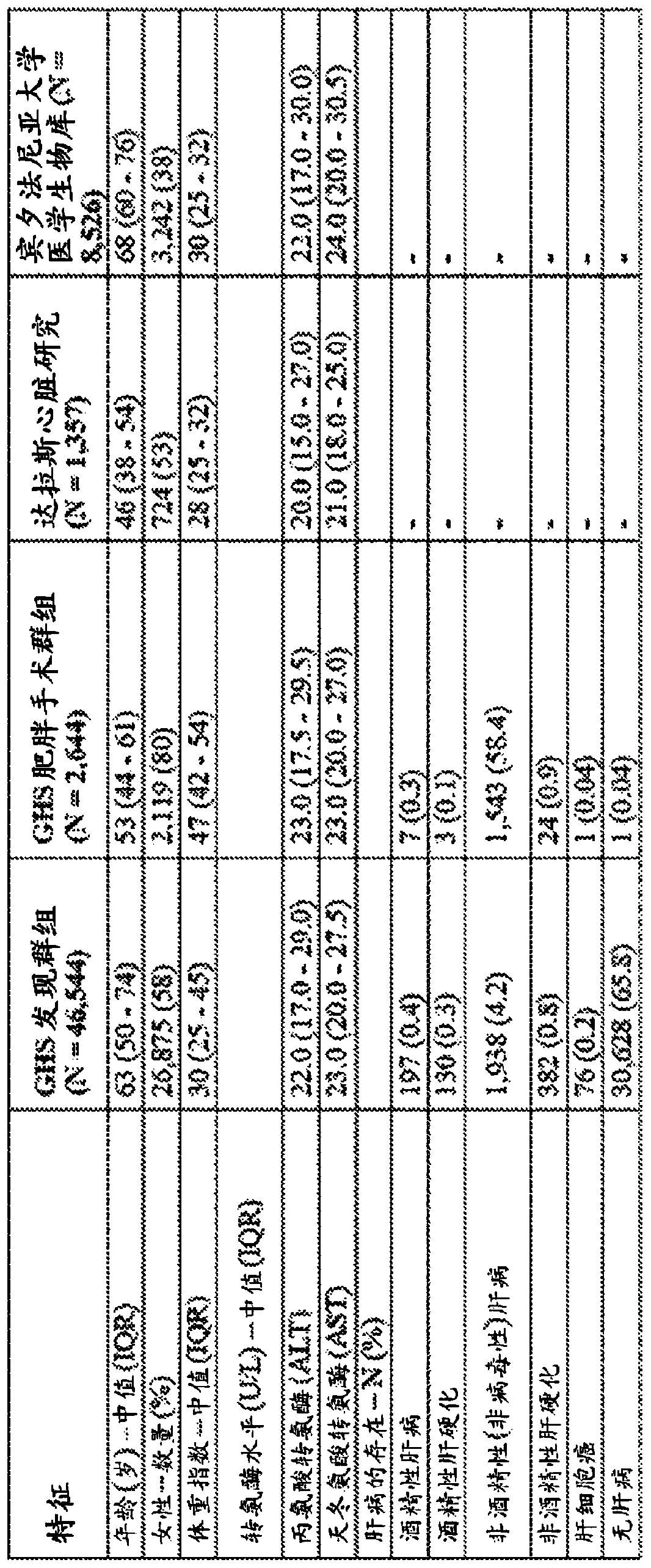
[0335] Human genetics studies were performed using genomic DNA samples and data from six cohorts. These studies included two Regeneron Genetics Center and Geisinger Health System (GHS) DiscovEHR research populations derived from the GHS20 The first 50,726 adult consenti...
Embodiment 2
[0374] Example 2: Gene expression analysis of HSD17B13 and PNPLA3 in 66 human liver samples
[0375] Gene expression of HSD17B13 and PNPLA3 was analyzed with 66 human liver samples. All samples were from control donors without steatosis, lobular inflammation, or fibrosis. The distribution of HSD17B13 rs72613567 (T / T, T / TA and TA / TA) and PNPLA3 rs738409 (C / C, C / G and G / G) genotypes is shown in Table 1.
[0376] genotype C / C C / G G / G ND T / T 12 8 1 0 T / TA 15 12 0 2 TA / TA 12 4 0 0
[0377] Expression of PNPLA3 was significantly reduced in homozygous alternate carriers of the HSD17B13 rs72613567 splice variant (see Figure 7 ). mRNA expression is shown in FPKM units. A 1.6-fold decrease compared to T / T with FDR 0.0071 was observed. The expression of the variant PNPLA3 C / C vector with the HSD17B13 TA / TA genotype was significantly reduced when compared to the HSD17B13 T / T vector: 1.7-fold decrease (FDR 0.017). Variant PNPLA3 C / G v...
Embodiment 3
[0380] Example 3: Gene expression analysis of HSD17B13 and PNPLA3 in 66 human liver samples
[0381] Association of exonic variants with aspartate and alanine aminotransferases
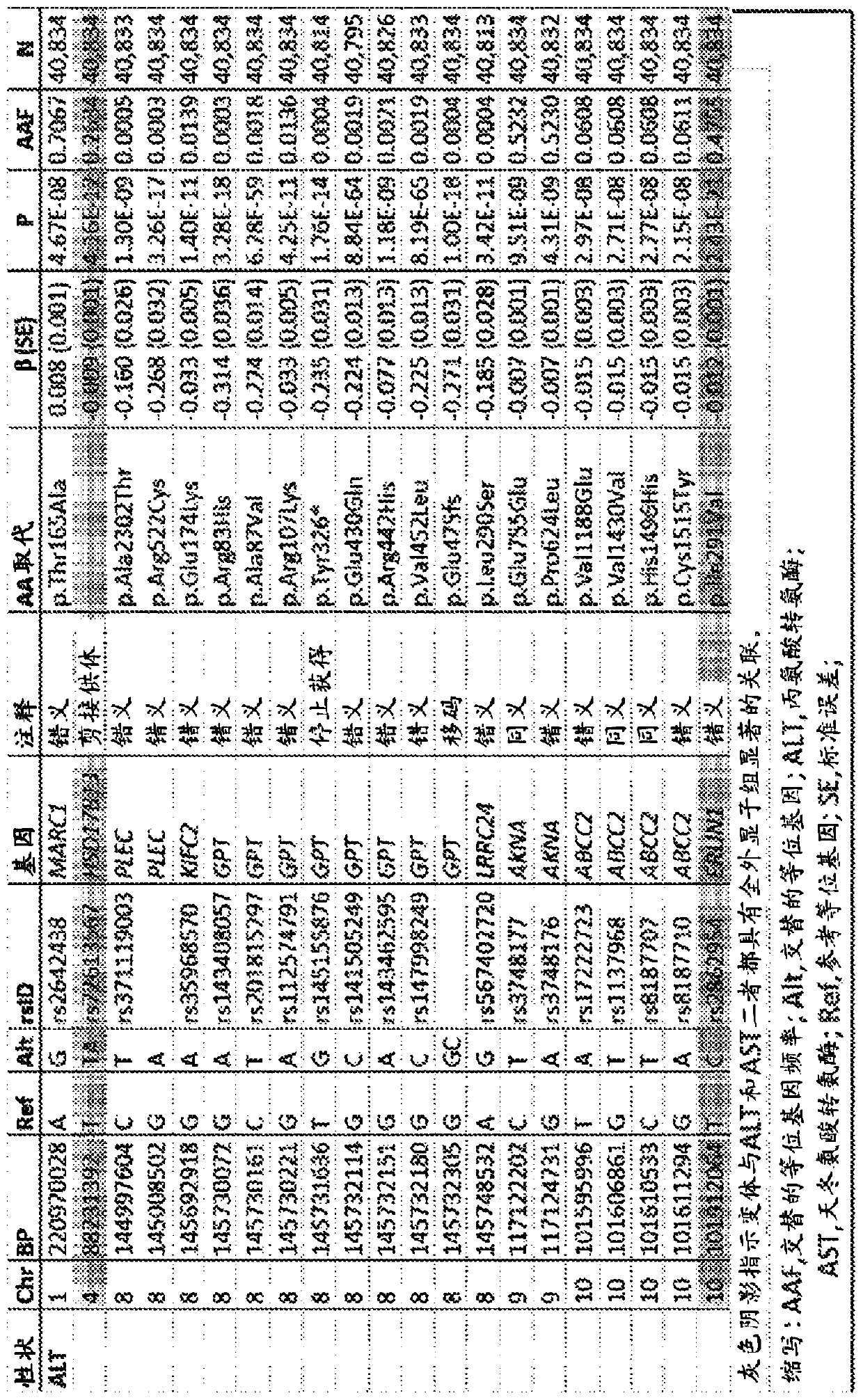
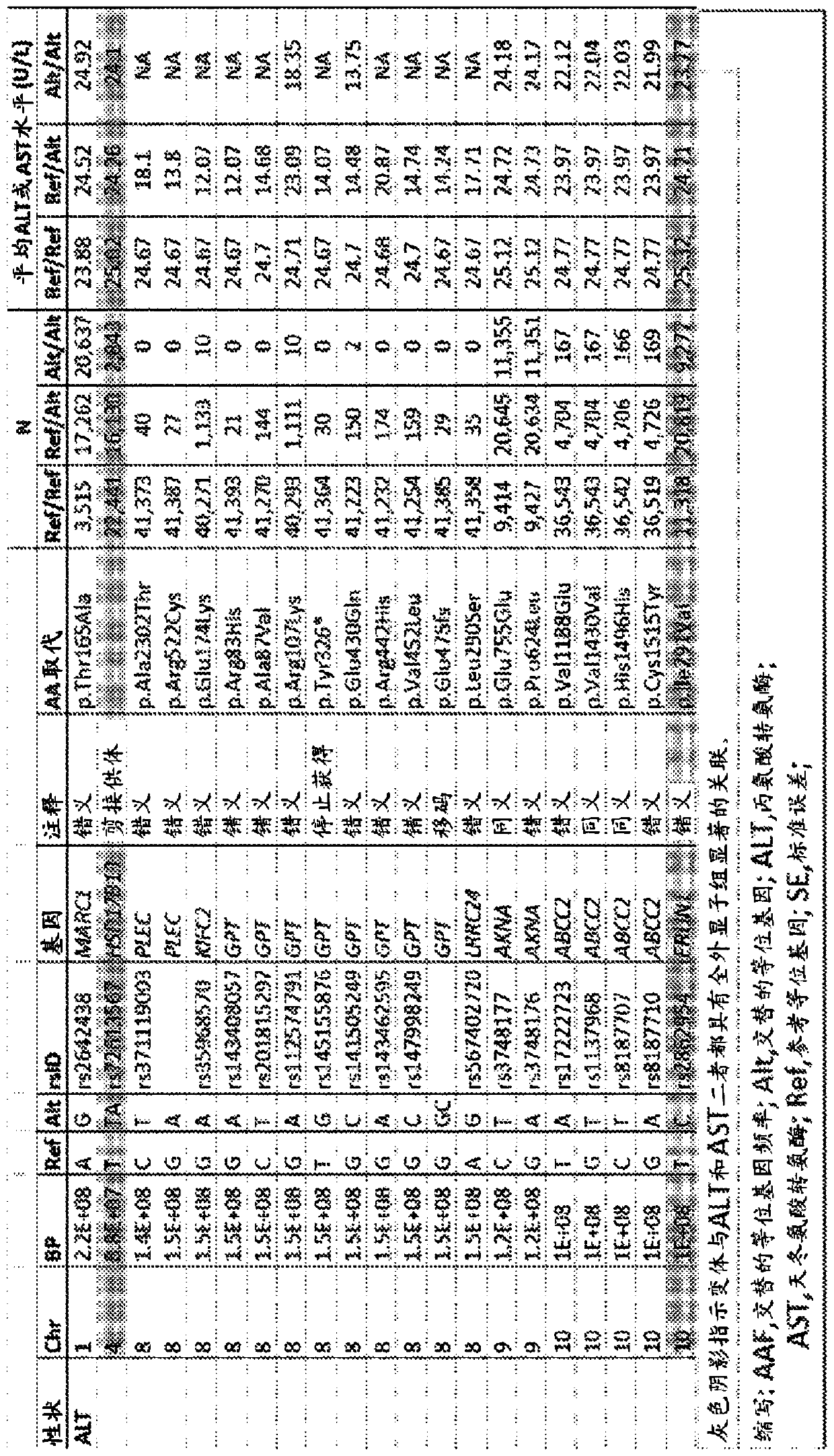
[0382] In a study from DiscovEHR (“GHS Discovery Cohort”; figure 1 The association of 502,219 biallelic single genetic variants with serum ALT or AST levels was examined in 46,544 individuals of European ancestry in Base Demographics in . A total of 35 variants in 19 genes were found with P-7 Related to ALT or AST (see Figure 14 and figure 2 ). see Figure 14 , shows a Manhattan plot (left) and a quantile-quantile plot (right) of the association of single nucleotide variants with serum transaminase levels in the GHS discovery cohort. 31 variants in 16 genes with P-7 Significantly correlated with alanine aminotransferase (ALT) levels (see panel A). 12 variants in 10 genes with P-7 Significantly correlated with aspartate aminotransferase (AST) levels (see panel B). All significant associations ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap