Methods and systems for diagnosis, prognosis and selection of treatment of leukemia
A technology for leukemia and prognosis, which is applied in the fields of disease diagnosis, biochemical equipment and methods, and the determination/examination of microorganisms, and can solve problems such as unexplainable
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
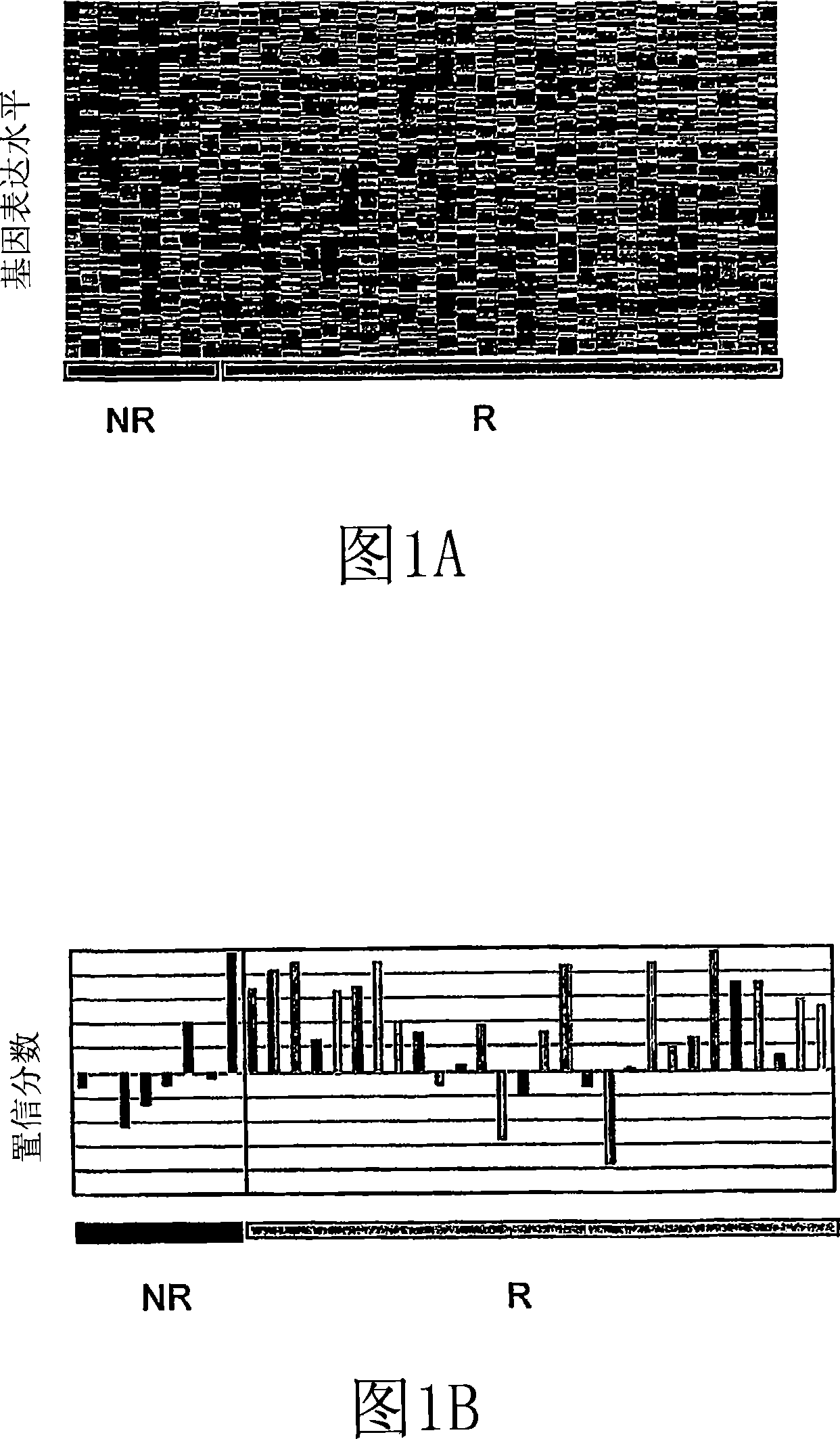
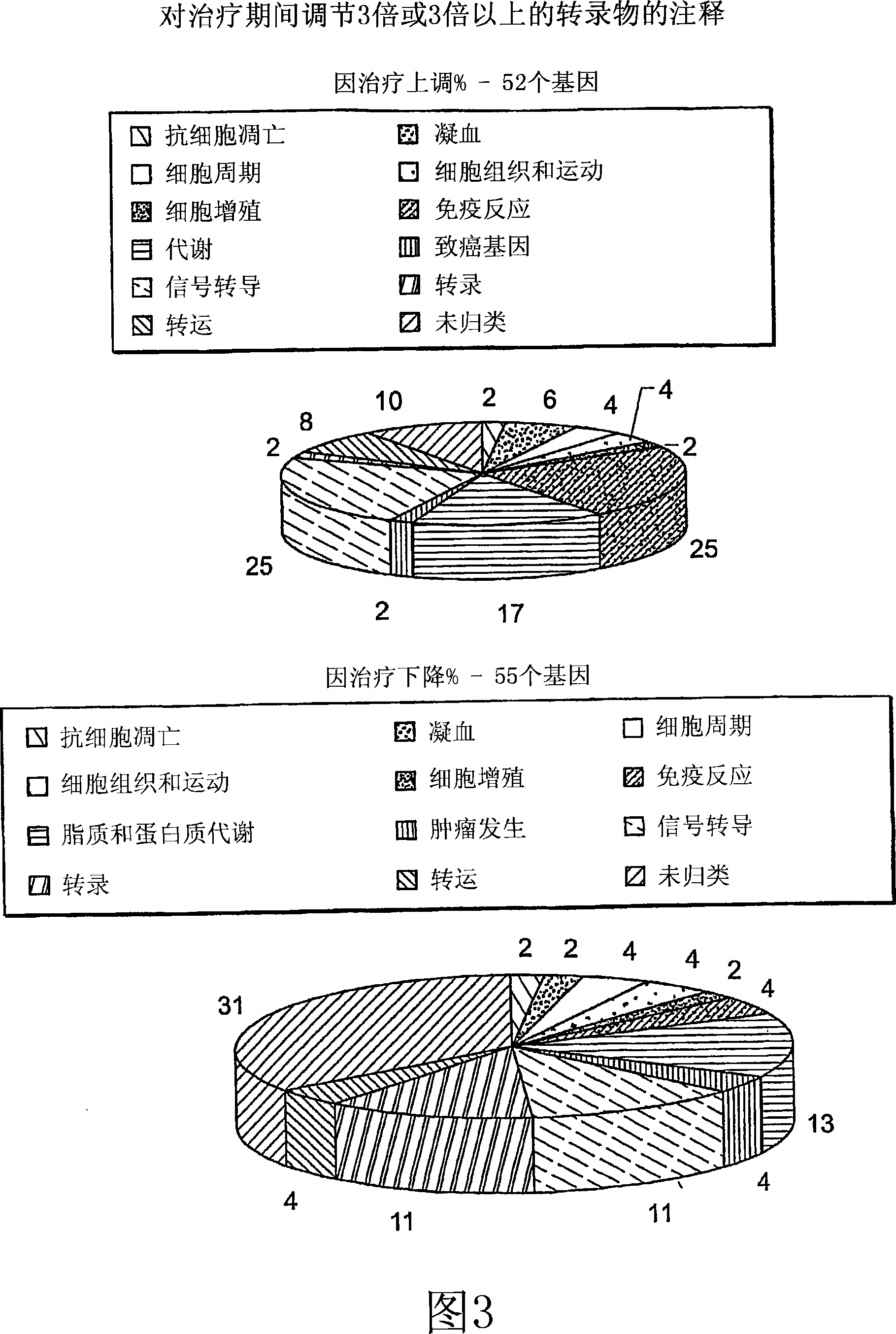
[0283] The above methods (including preparation of blood samples, assembly of class predictors, and construction and comparison of expression profiles) can be easily used to diagnose or monitor the development, progression or treatment of AML. This can be achieved by comparing the expression profile of one or more AML disease genes in the individual of interest with a reference expression profile of at least one AML disease gene. The reference expression profile may include an average expression profile or a group of individual expression profiles, each of which represents the expression of peripheral blood genes in a specific AML patient or disease-free human. The similarity between the expression profile of the individual of interest and the reference expression profile indicates the presence or absence of the AML disease state. In many embodiments, the disease genes used for AML diagnosis are selected from Table 7.
[0284] One or more AML disease genes selected from Table 7 ca...
example 1
[0297] Example 1. Clinical trials and data collection
[0298] experimental design
[0299] The AML patients (13 women and 23 men) were all white and had an average age of 45 years (in the range of 19-66 years). The inclusion criteria for AML patients include a 20% excess of blast cells in the bone marrow; morphological diagnosis of AML according to the FAB classification system; and flow cytometry that indicates a positive CD33+ status. Participation in clinical trials requires a consistent pathological diagnosis of AML by the on-site pathologist following the histological evaluation of bone marrow aspiration. A summary of the patient's cytogenetic characteristics is presented in Table 11.
[0300] Cytogenetic characteristics of PG consenting AML patients in Table 11.0903B1-206-US contributing to the baseline sample
[0301] Cytogenetic characteristics
PG agrees (n=36) *
Normal karyotype
12(33%)
Compound karyotype (>3 abnormalities)
6(1...
example 2
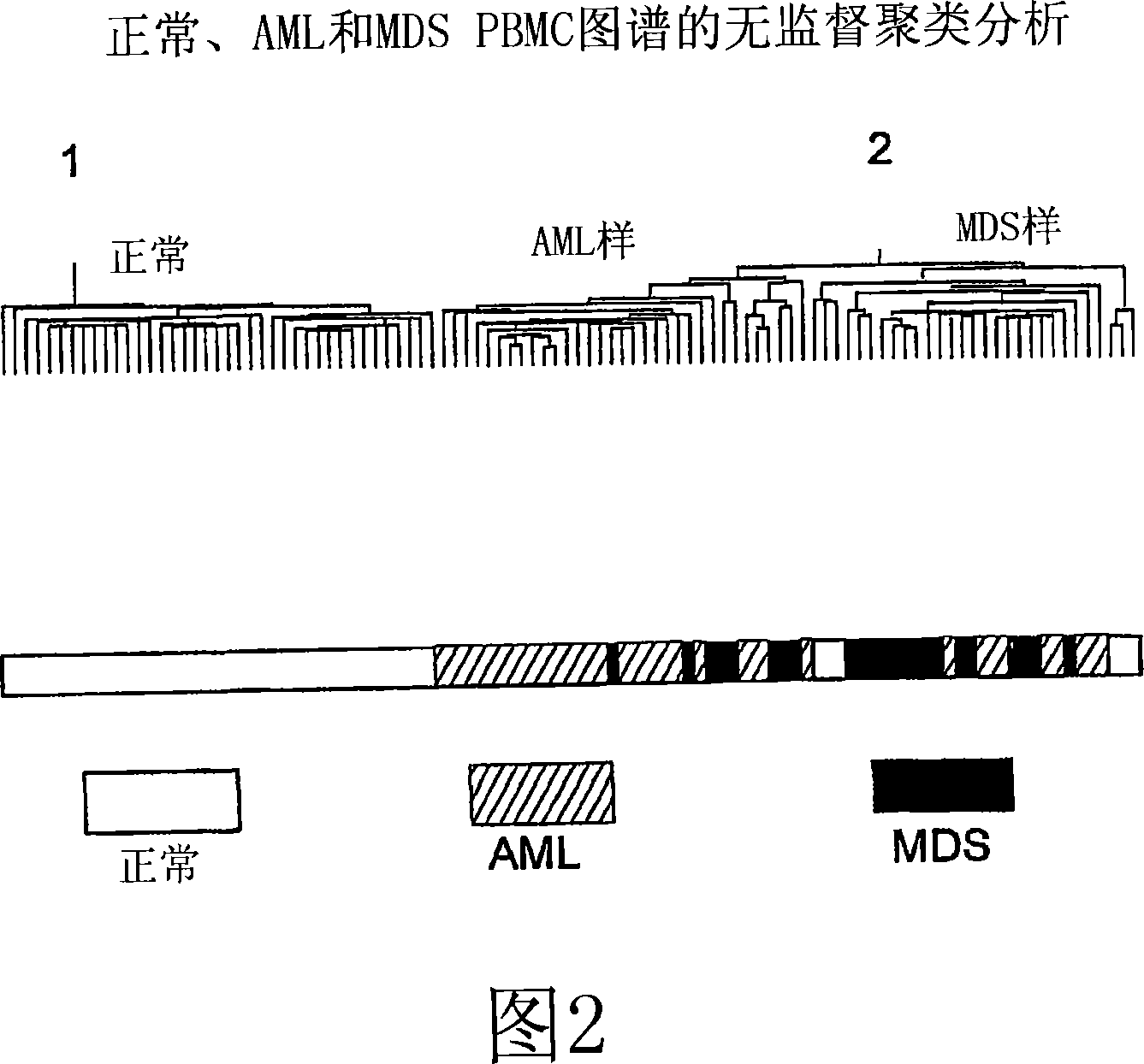
[0311] Example 2. Disease-related transcripts in AML PBMC
[0312] The scaled frequency normalization method was used to standardize 36 AML PBMC samples with the U133A transcription profiles of 20 MDS PBMC and 45 healthy volunteers PBMC. A total of 7879 transcripts in one or more patterns with a maximum frequency greater than or equal to 10 ppm (expressed as 1P, 1≥10 ppm) in the pattern are detected.
[0313] To identify AML-related transcripts, the average fold difference between AML and normal PBMC was calculated by dividing the average expression level in the AML profile by the average expression level in the normal profile. Student's t test (two samples, unequal variance) was used to evaluate the significance of expression differences between groups.
[0314] For unsupervised hierarchical clustering, 7879 transcripts satisfying the expression filter factor 1P, 1≥10ppm were used. The data was log-transformed, and the gene expression values were median-centered, and the aver...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com