Parallel universal sequence alignment method running on multi-core computer platform
A multi-core computer and general-purpose sequence technology, applied in computing, special data processing applications, instruments, etc., can solve the problem of low efficiency of sequence alignment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
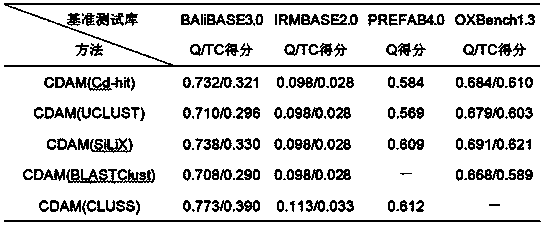
[0086] The experiment tested two sets of data respectively. One set was the traditional sequence comparison Benchmarks, including BAliBASE3.0, IRMBASE2.0, PREFAB4.0 and OXBench1.3, which were used to calculate the Q / TC score of the CDAM method to evaluate its Alignment accuracy. One group uses the Rose sequence generator to generate a large-scale sequence collection, which is used to calculate the speedup ratio of the CDAM method and the MUSCLE method, so as to evaluate the efficiency of the CDAM method in processing large-scale sequence alignments.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com