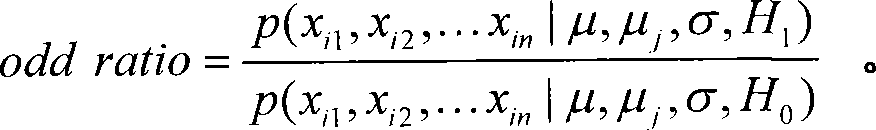Method for screening gene chip difference expression gene
A technology for differentially expressed genes and screening genes, applied in the field of differentially expressed genes screening, which can solve problems such as large experimental errors, difficulty in estimating algorithm sensitivity and specificity, and lack of analytical methods for chip data.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
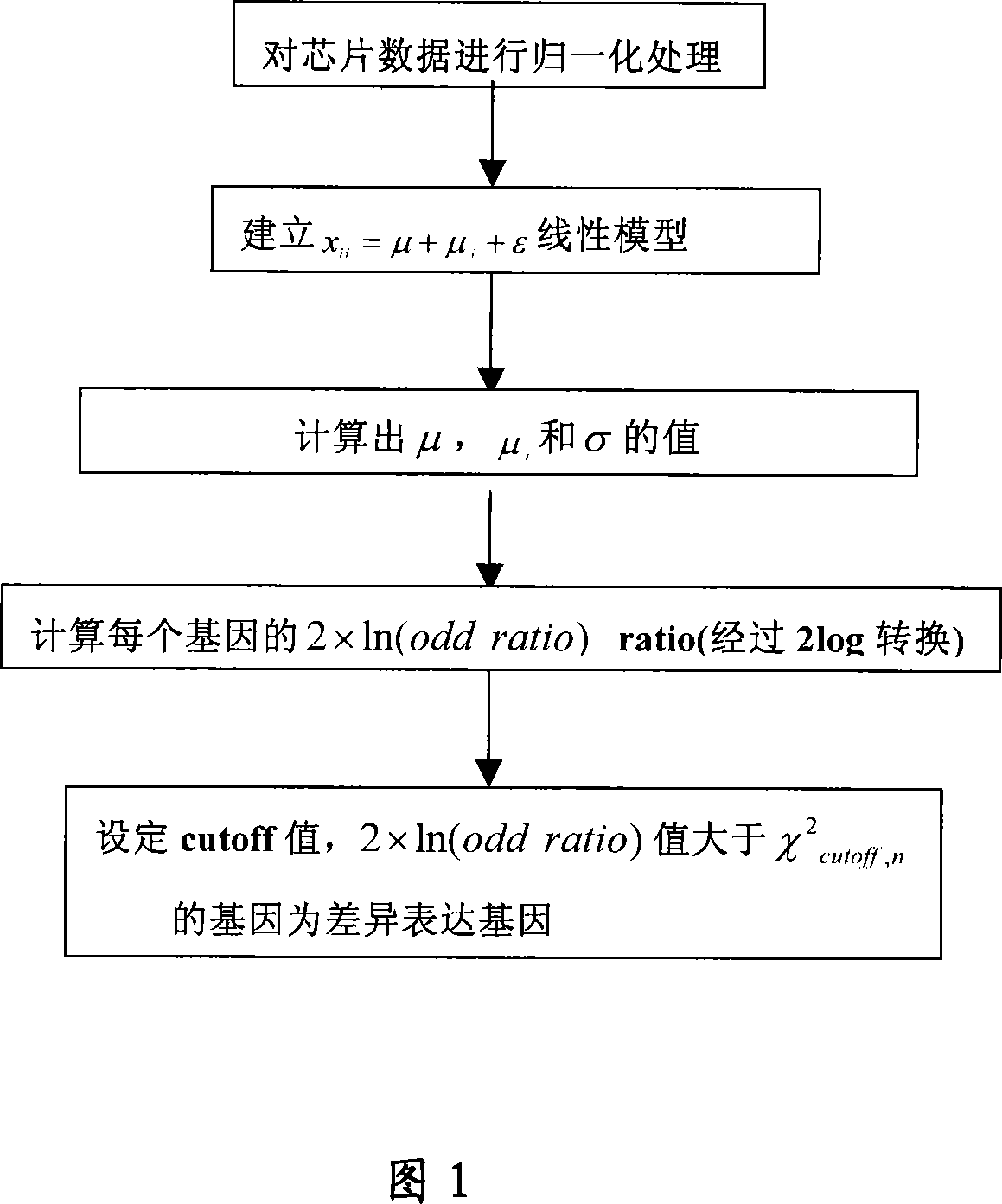
[0010] The specific method is described as follows:
[0011] First, use chip scanning image processing software (such as GenePix pro 4.0) to obtain expression value data at the gene level. Then, normalize the chip data between chips. Signal values from ChIP results were then converted into ratios relative to control experiments. Take the logarithm of the ratio (it is better to take e as the base). We used this log ratio (ln ratio) as the basis for the analysis.
[0012] Suppose we have n gene chips (corresponding to n samples, typically, 1
[0013]
[0014] where x ij is the ln ratio value of the i-th (1≤i≤m) gene in the j-th (1≤j≤n) chip.
[0015] Next we build a linear model:
[0016] x ij =μ+μ j +ε ②
[0017] where μ is the global mean, μ j is the column effect, and ε is the residual. We assume ε~N(0,σ 2 ). That is, it is assumed that the residual ε in different chips conforms to a norm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com