Gene base editor
An editor, base technology, applied in chemical instruments and methods, antibody mimics/scaffolds, hybrid peptides, etc., can solve the problem of limiting the effective editing sites of base editors, unable to effectively correct, unable to effectively edit bases Base C, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0084] Example 1: Base editor
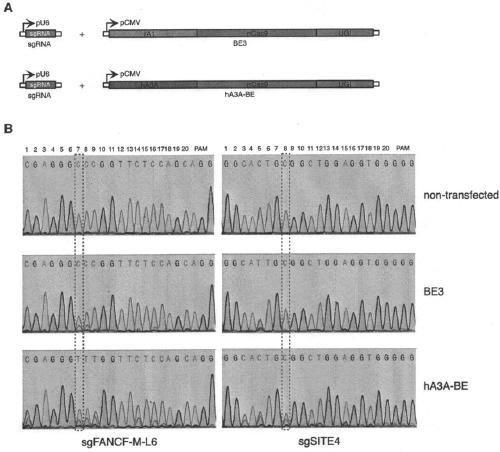
[0085] The expression plasmid of pCMV-hA3A-BE was constructed. Human apolipoprotein B messenger RNA deaminase catalytic subunit 3A (APOBEC3A, hA3A; SEQ ID NO: 1) with Cas9 nickase and a uracil DNA glycosidase inhibitor [Bacillus phage] (SEQ ID NO: 12 ) fused to an expression vector. The 10th aspartic acid of Cas9 nickase is mutated to alanine, thereby losing the activity of cutting double strands and ensuring a nick on one strand.
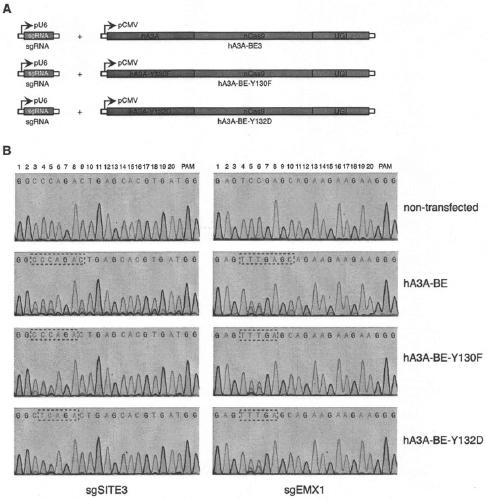
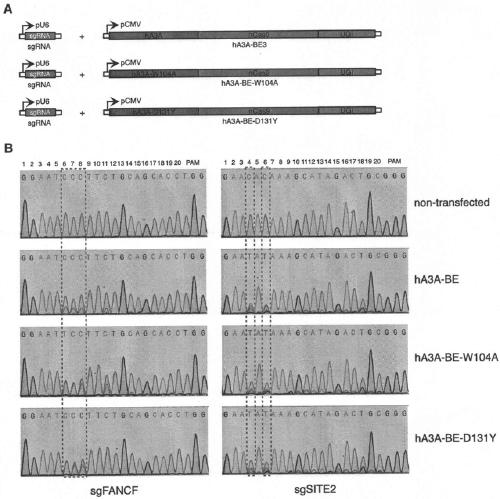
[0086] The fusion expression vector hA3A-nCas9-UGI (hA3A-BE, SEQ ID NO: 21) and the expression vector of single-stranded guide RNA were co-transformed into eukaryotic cells ( figure 1 , legend A), C-T base editing occurs at the site targeted by the guide RNA of the genome. The sequence of the genomic DNA target position was amplified by PCR, and the base editing efficiency of the target site C-T was detected by Sanger DNA sequencing. Compared with the co-expression of sgRNA and BE3, the method of co-expression of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com