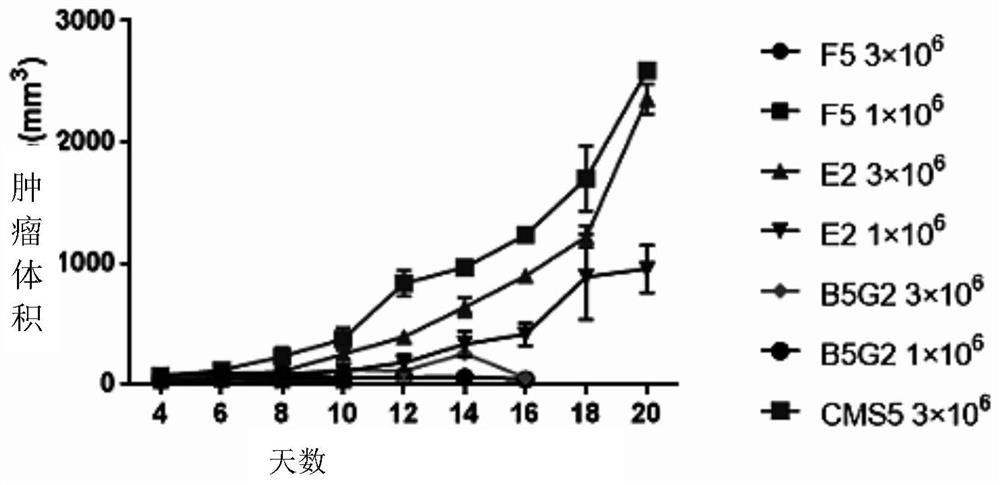
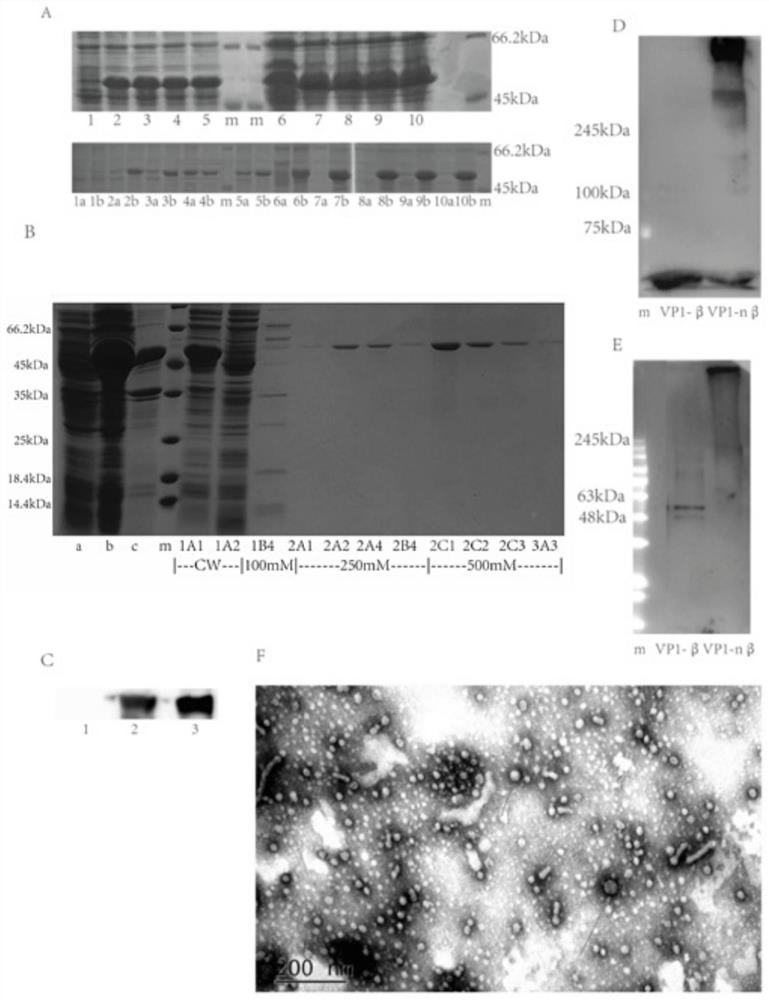
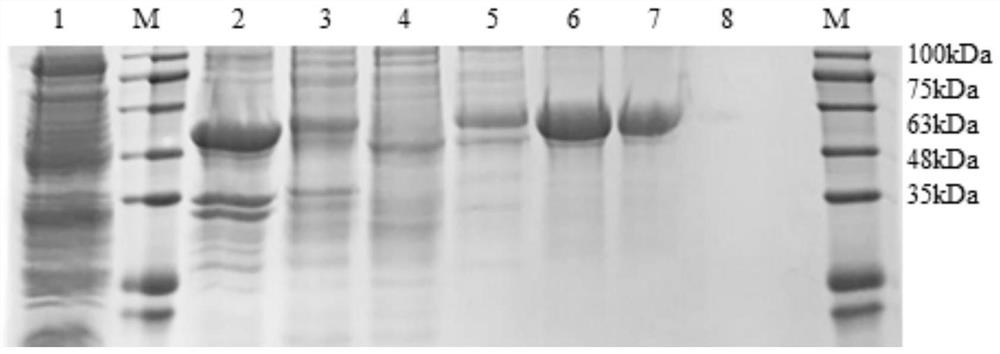
A novel vaccine for the prevention and treatment of Merkel cell carcinoma
A cell carcinoma, Merkel's technology, applied in the fields of immunology and virology, can solve problems such as poor immune effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
[0088] Basic experimental example 1 Construction, screening and verification of CMS5-VP1 tumor cell line
[0089] The CMS5-VP1 tumor cell line will be mainly used for the verification of vaccine efficacy.
[0090] 1. Construction of pcDH-VP1 plasmid
[0091] (1) First, design primers according to the pcDH-GFP plasmid:
[0092] MCV-F(5'-CTAGAGCTAGCGTTTAAACTTAAGC-3');
[0093] MCV-R (5'-GCGGCCGCTCTAGACTCGAGTCACAGCTCCTG-3').
[0094] The 5' end of MCV-F contains a Nhe I restriction site, and the 3' end of MCV-R contains a Not I restriction site.
[0095] (2) PCR amplification
[0096] Using plasmid pVAX-VP1 as a template, a VP1 fragment containing an endonuclease site at the N-terminus and C-terminus was obtained by amplification. The PCR reaction system is shown in Table 1 below.
[0097] Table 1 PCR reaction reagent dosage table
[0098]
[0099] The PCR reaction program is shown in Table 2 below.
[0100] Table 2 PCR reaction program
[0101]
[0102] (3) Enzyme...
experiment example 2
[0225] Basic experimental example 2MCV-VP1_VP2 pseudovirus construction, expression verification
[0226] 1. Plasmid construction
[0227] The plasmid pcDNA3.1-VP2 was synthesized by Nanjing GenScript, inserted into the multi-cloning site of vector pcDNA3.1 by endonuclease Kpn I / Xho I, and the correctness of the insertion site was confirmed by sequencing.
[0228] The pVAX1-VP1 / DH5α and pcDNA3.1-VP2 / DH5α glycerol bacteria were streaked on LB plates (containing corresponding antibiotics such as Amp or Kan), and cultured in a 37°C incubator overnight.
[0229] The pVAX1-VP1 / DH5α and pcDNA3.1-VP2 / DH5α monoclonal colonies were respectively picked and placed in 5 mL LB liquid medium (containing corresponding antibiotics such as Amp or Kan), and incubated overnight at 37°C with shaking on a shaker.
[0230] The plasmids of pVAX1-VP1 / DH5α and pcDNA3.1-VP2 / DH5α cultured overnight were extracted with plasmid mini-suction kit.
[0231] Using plasmid pcDNA3.1-VP2 as the carrier and pVA...
Embodiment 1
[0273] Example 1 A DNA vaccine against Merkel cell carcinoma
[0274] The nucleotide sequence shown in SEQ ID NO. 1 was cloned into the pVAX1 vector, and the enzyme cleavage sites used for the cloning were XbaI and HindIII.
[0275] The specific steps of cloning are conventional methods in the art, which will not be repeated here. Cloning methods include PCR amplification, restriction digestion, ligation and related steps.
[0276] The recombinant vector obtained by successfully cloning the nucleotide sequence shown in SEQ ID NO. 1 into the pVAX1 vector was named pVAX1-MCV-VP1 according to the conventional naming convention in the art.
[0277] The identification method is a conventional method in the field, and will not be repeated here. For example, double-enzyme digestion identification or sequencing identification. After double digestion with XbaI and HindIII, a DNA band of about 1.3kb can be observed.
[0278]pVAX1-MCV-VP1 was amplified using conventional kits in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com