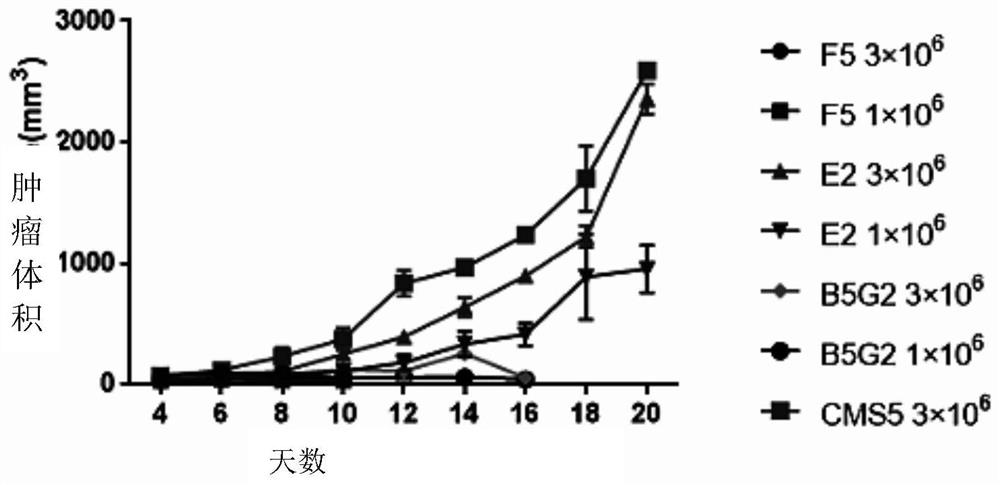
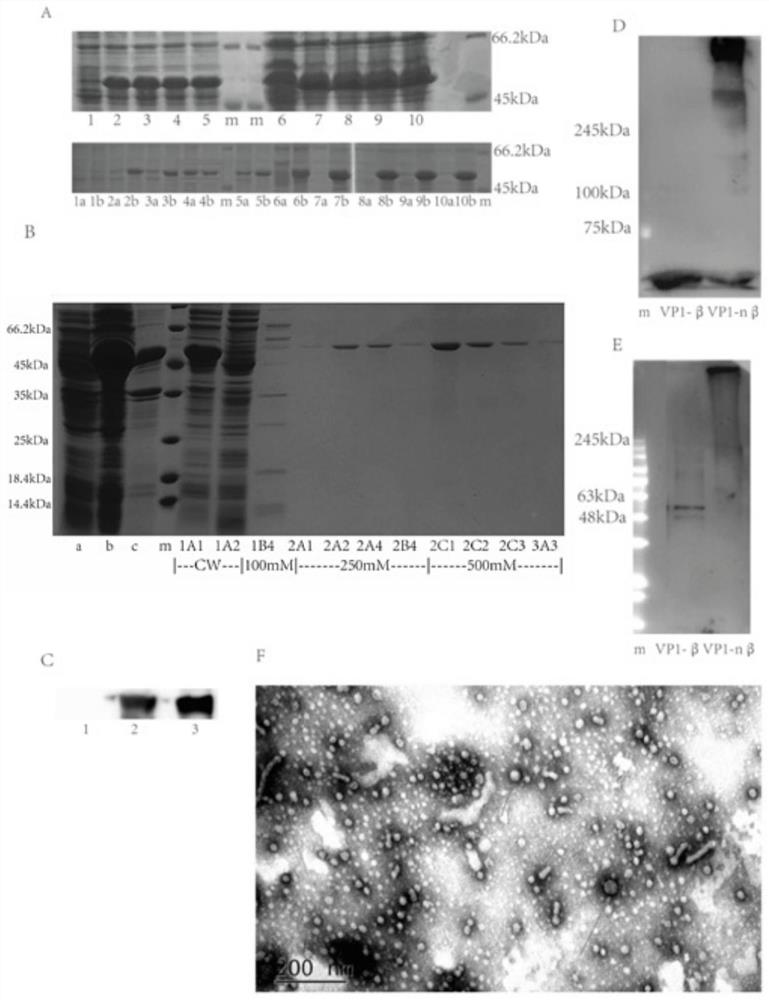
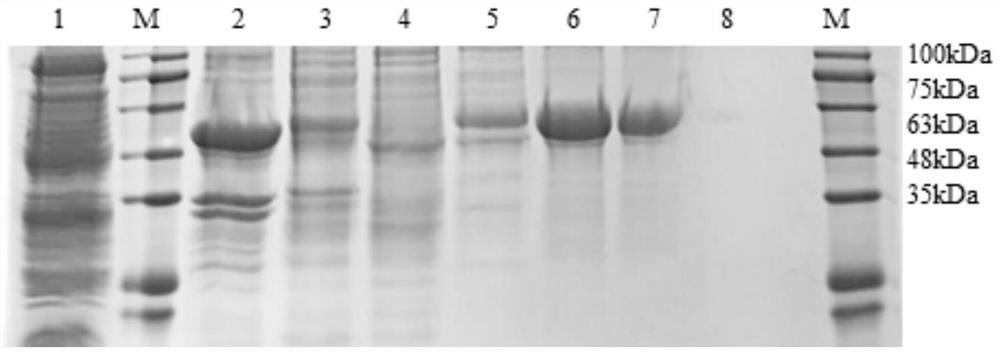
Novel vaccine for preventing and treating Merkel cell carcinoma
A cell cancer, Merkel's technology, applied in the field of immunology and virology, can solve the problem of poor immune effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
experiment example 1
[0088] Basic experiment example 1 Construction, screening and verification of CMS5-VP1 tumor cell line
[0089] The CMS5-VP1 tumor cell line will be mainly used in the verification of the vaccine effect.
[0090] 1. Construction of pcDH-VP1 plasmid
[0091] (1) First, design primers according to the pcDH-GFP plasmid:
[0092] MCV-F (5'-CTAGAGCTAGCGTTTAAACTTAAGC-3');
[0093] MCV-R (5'-GCGGCCGCTCTAGACTCGAGTCACAGCTCCTG-3').
[0094] The 5' end of MCV-F contains a Nhe I restriction site, and the 3' end of MCV-R contains a Not I restriction site.
[0095] (2) PCR amplification
[0096] Using the plasmid pVAX-VP1 as a template, VP1 fragments containing endonuclease sites at the N-terminus and C-terminus were amplified respectively. The PCR reaction system is shown in Table 1 below.
[0097] Table 1 Amount of PCR reaction reagents
[0098]
[0099] The PCR reaction program is shown in Table 2 below.
[0100] Table 2 PCR reaction program
[0101]
[0102] (3) enzyme di...
experiment example 2
[0225] Basic experiment example 2 MCV-VP1_VP2 pseudovirus construction, expression verification
[0226] 1. Plasmid construction
[0227] Plasmid pcDNA3.1-VP2 was synthesized by Nanjing GenScript, inserted into the multiple cloning site of vector pcDNA3.1 by endonuclease Kpn I / Xho I, and the correctness of the insertion site was confirmed by sequencing.
[0228]Streak the pVAX1-VP1 / DH5α and pcDNA3.1-VP2 / DH5α glycerol bacteria on an LB plate (containing corresponding antibiotics such as Amp or Kan) and cultivate overnight in a 37°C incubator.
[0229] Single clone colonies of pVAX1-VP1 / DH5α and pcDNA3.1-VP2 / DH5α were respectively picked and placed in 5 mL LB (containing corresponding antibiotics such as Amp or Kan) liquid medium, and cultured on a shaking table at 37°C overnight.
[0230] Plasmids were extracted from pVAX1-VP1 / DH5α and pcDNA3.1-VP2 / DH5α cultured overnight with a small plasmid extraction kit.
[0231] Using the plasmid pcDNA3.1-VP2 as the vector and pVAX1-VP1 ...
Embodiment 1
[0273] Example 1 A kind of DNA vaccine against Merkel cell carcinoma
[0274] The nucleotide sequence shown in SEQ ID NO.1 was cloned into the pVAX1 vector, and the restriction sites used for cloning were XbaI and HindIII.
[0275] The specific steps of cloning are conventional methods in the art, and will not be repeated here. The cloning method includes PCR amplification, enzyme cutting, connection and related steps.
[0276] The recombinant vector obtained by successfully cloning the nucleotide sequence shown in SEQ ID NO.1 into the pVAX1 vector was named pVAX1-MCV-VP1 according to conventional naming conventions in the art.
[0277] The method of identification is a conventional method in the art, and will not be repeated here. For example, double enzyme digestion identification or sequencing identification. After double digestion with XbaI and HindIII, a DNA band of about 1.3kb can be observed.
[0278] pVAX1-MCV-VP1 was amplified using a conventional kit in the art, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com