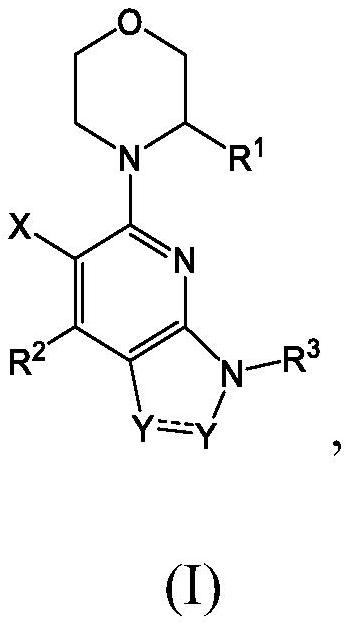
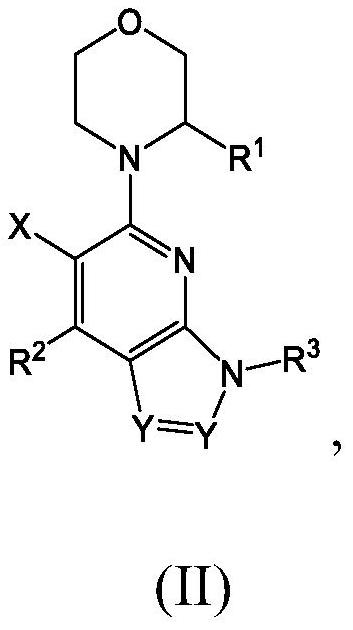
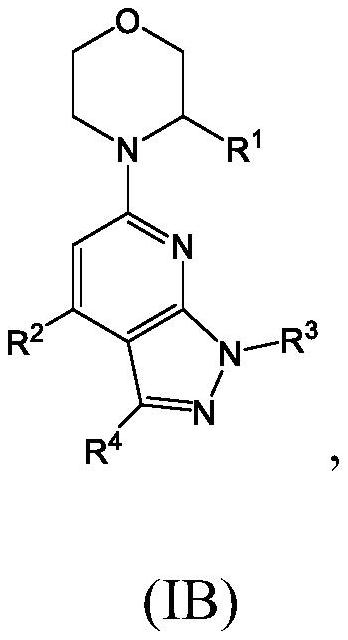
Compounds, pharmaceutical compositions, and methods of preparing compounds and of their use as atr kinase inhibitors
A compound and pharmaceutical technology, applied in the field of compounds and pharmaceutical compositions, can solve the problems of rapidly proliferating tissues and stem cell populations
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0719] Embodiment 1. The preparation of compound
[0720] Compound 1
[0721] Step 1. Use vacuum / N 2 A suspension of 4-chloro-7-azaindole (25 g) in DMA (140 mL) was gas sparged (3 cycles). Then add zinc powder (1.07g), zinc cyanide (11.26g), dppf (2.72g) and Pd 2 (dba) 3 (2.39g). Vacuum again / N 2 The mixture was gas-swept (3 cycles) and heated to 120 °C for 4 h. The reaction mixture was cooled to 100 °C and water (428 mL) was added within 30 min. The mixture was then cooled to rt within 2h. The crude product was filtered and washed with water (2 x 95 mL), then added to 3N HCl (150 mL), and the mixture was stirred at rt for 2 h. Insolubles were removed by filtration. To the filtrate was added 50% aqueous NaOH until pH 12 was reached. Filtration and drying afforded 1H-pyrrolo[2,3-b]pyridine-4-carbonitrile (11.6 g) as a tan solid.
[0722] Step 2. A mixture of 1H-pyrrolo[2,3-b]pyridine-4-carbonitrile (10.4 g) and NaOH (29 g) in water (100 mL) and EtOH (100 mL) was heat...
Embodiment 2
[0839] Embodiment 2.ATR / ATRIP enzymatic assay
[0840] Detection of ATR kinase activity The AlphaScreen system was used to measure the phosphorylation of the substrate protein p53. In assay buffer (50mM Hepes pH 7.4, 0.1mM vanadate, 0.5mM DTT, 0.1mM EGTA, 5mM MnCl 2 , 0.01% Brij-30, 1% Glycerol, 0.05% BSA) recombinant purified ATR / ATRIP (Eurofins cat. no. 14-953) at a final concentration of 0.63 nM was mixed with serially diluted compounds in 10% DMSO. The final DMSO concentration was 1.25%. A premix of GST-tagged p53 (full-length, Enzo Life Sciences cat. no. BML-FW9370) and 5'-adenosine triphosphate (Sigma-Aldrich cat. no. 10519979001, Roche Diagnostic) in assay buffer was added to the enzyme:compound In the mixture, the final concentration was 25 nM GST-p53 and 3 μM ATP. The reaction was allowed to proceed for 1 hour at room temperature, and was then reacted by adding a final dilution of 1:3000 phosphorylated p53 (Ser 15) antibody (New England Biolabs cat# 9284S), buffer ...
Embodiment 3
[0841] Example 3. ATR assay in Hela cells
[0842] HeLa S3 cells were plated in a 384-well format at a density of 16K cells per 25 μL well in regular medium F-12K 10% FBS, and incubated at 37 °C, 5% CO 2 Incubate overnight. Then, the medium was replaced by 20 μL Opti-MEM (without phenol red) per well, and 5 μL of serially diluted compounds were added to the assay plate at a final concentration of 0.5% DMSO. Cells and compounds were incubated at room temperature for 20 min before adding 5 μL of gemcitabine at a final concentration of 1.5 μM. Incubate the plate at 37 °C, 5% CO 2 Incubate for 3.5 to 4 hours. Media was removed and cells were lysed in 15 μL of PerkinElmer Lysis Buffer for 10-20 min; 4 μL of lysate was then transferred to 384 format proxi white plates (PerkinElmer Life Sciences cat# 6008280). Phosphorylation of CHK1 at Ser345 was quantified using Alphascreen SureFire CHK1 p-Ser345 (PerkinElmer Life Sciences Cat. No. TGRCHK1S10K) and Alphascreen protein A. (Perki...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com