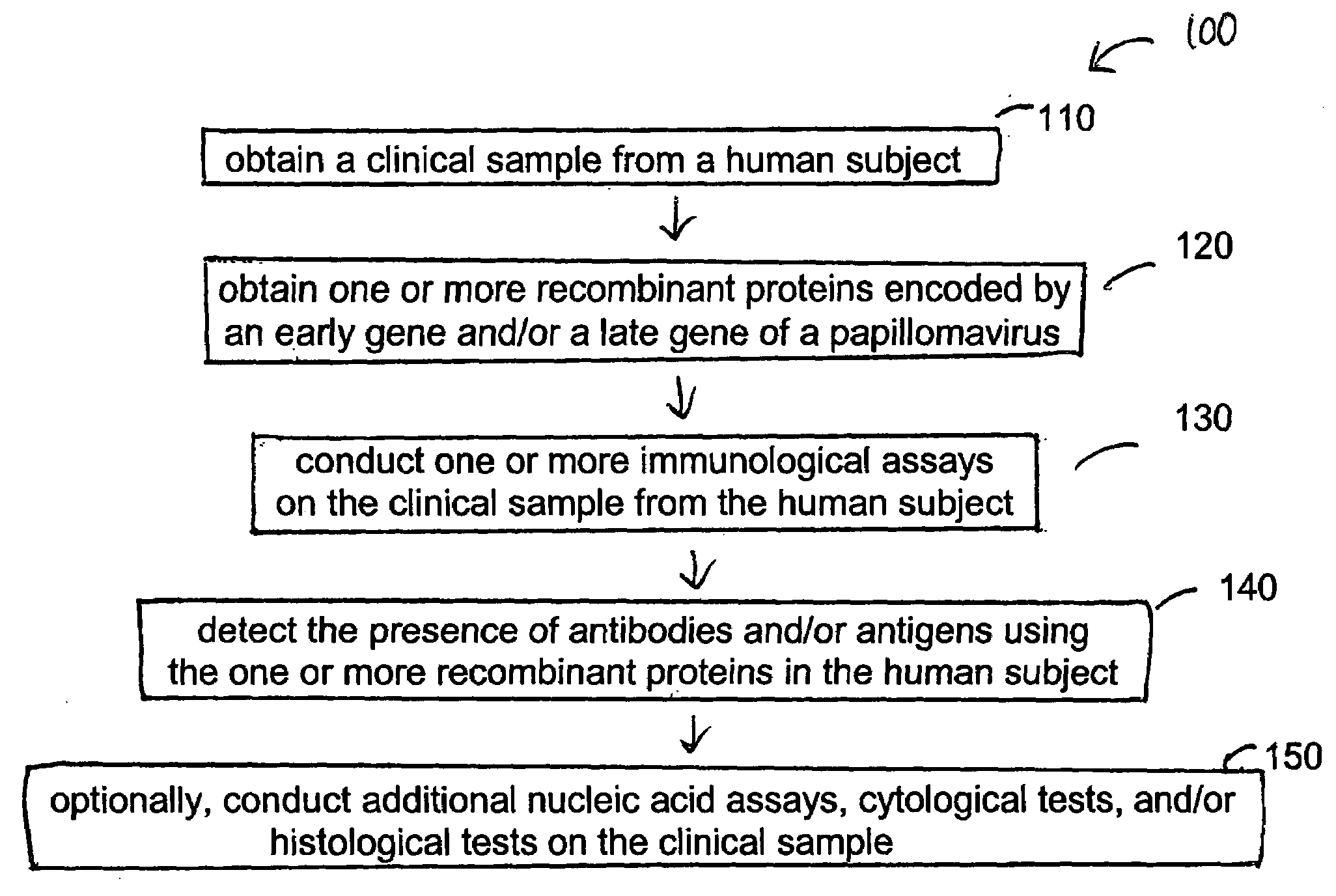
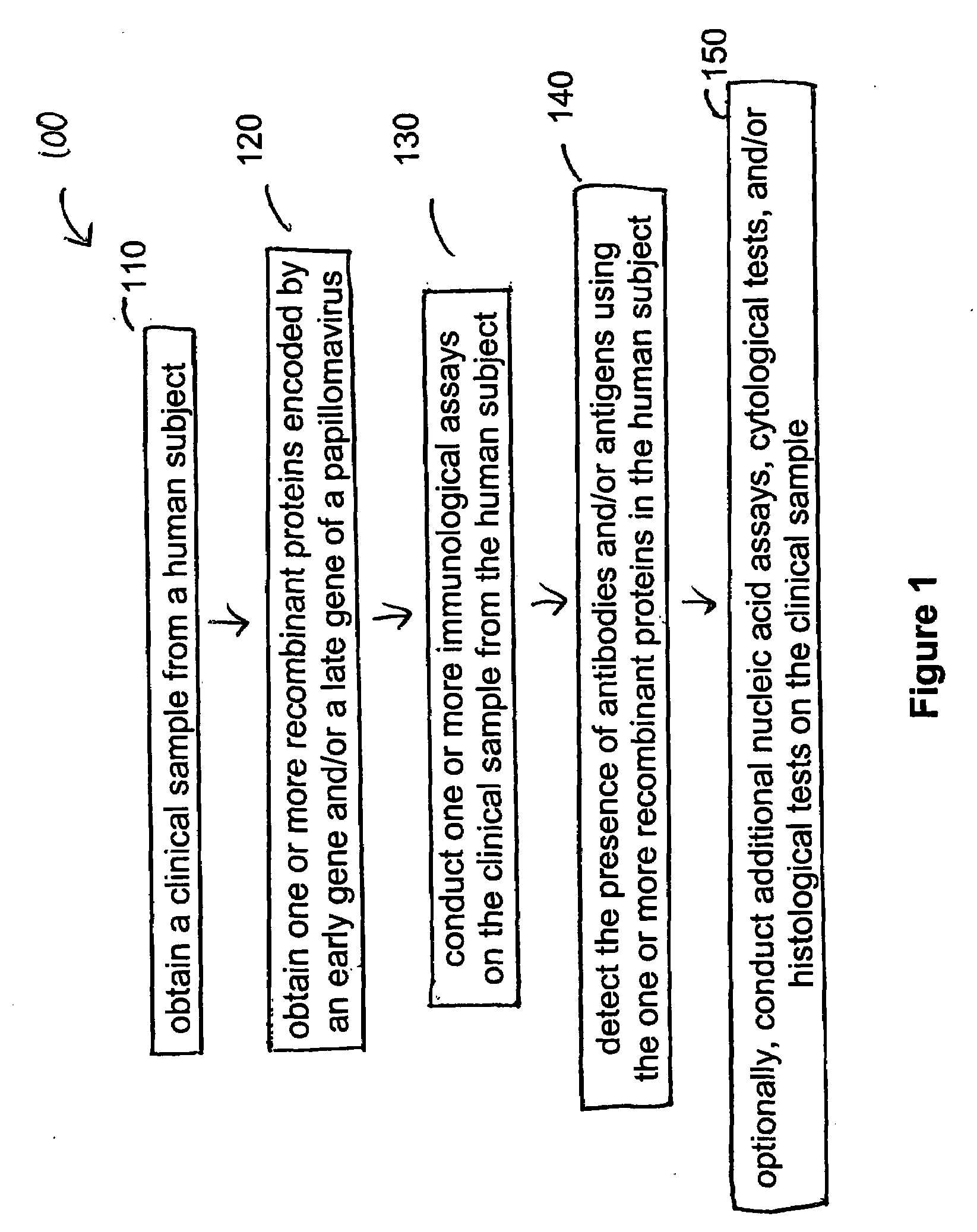
Detection method for human pappilomavirus (HPV) and its application in cervical cancer
a detection method and technology for human pappilomavirus, applied in the field of detection method for human pappilomavirus and its application in cervical cancer, can solve the problems of high risk of progression toward invasive cancer, regress spontaneously, and usually self-limited
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Cloning and Production of Various Recombinant Proteins Encoded by HPV-16, Early E6 Gene
[0088]Cloning of an exemplary oncogenic E6 early gene from an exemplary HPV type, HPV-16, is described herein. A 474 base pair (b.p.) DNA fragment (SEQ ID NO. 1) containing the 157 amino acid coding region (SEQ ID NO. 2) of the HPV-16 E6 gene was obtained by polymerase chain reaction (PCR) amplification. Primers were used for cloning, for example, a pair of forward and reverse primers, 5′ cgcGGATCCcaccaaaagagaactgcaatgtttc 3′ (SEQ ID NO. 3) and 5′ cccAAGCTTttacagctgggtttctctacgtg 3′ (SEQ ID NO. 4), respectively. The DNA sequence of the isolated DNA fragment was confirmed by comparing with the sequence from Gene Bank database. All cloning procedures are carried out according to the protocols described in “Molecular Cloning”, A Laboratory Manual, eds. Sambrook, Fritsch and Maniatis, Cold Spring Harbor Laboratory Press, 1989. In addition, E6 DNA fragments from different strains of HPV-16 can also be ...
example 2
Cloning and Production of Recombinant Proteins Encoded by HPV-16 Early E7 Gene
[0095]Cloning of an exemplary oncogenic E7 early gene from an exemplary HPV type, HPV-16, is described herein. A 294 base pair (b.p.) DNA fragment (SEQ ID NO. 7) containing the 99 amino acid coding region (SEQ ID NO. 8) of the HPV-16 E7 gene was obtained by polymerase chain reaction (PCR) amplification. Primers were used for cloning, for example, a pair of forward and reverse primers, 5′ cgcGGATCCcatggagatacacctacattgc 3′ (SEQ ID NO. 9) and 5′ ccgGAATCttatggtttctgagaacagatgg 3′ (SEQ ID NO. 10), respectively. The DNA sequence of the isolated DNA fragment was confirmed by comparing with the sequence from Gene Bank database. In addition, E7 DNA fragments from different strains of HPV-16 can also be cloned from different clinical samples or sources.
[0096]The obtained 294 base pair (b.p.) DNA fragment was sub-cloned into a GST expression vector in order to express a recombinant HPV-16 E7 GST fusion protein. The...
example 3
Cloning and Production of Recombinant Proteins Encoded by HPV-16 Late L1 Gene
[0098]Cloning of an exemplary late gene from an exemplary HPV type, HPV-16, is described herein. A 1596 base pair (b.p.) DNA fragment (SEQ ID NO. 13) containing the 531 amino acid coding region (SEQ ID NO. 14) of the HPV-16 L1 gene was obtained by polymerase chain reaction (PCR) amplification. Primers were used for PCR cloning, for example, a pair of forward and reverse primers, 5′ ccgCTCGAGatgcaggtgacttttatttacatcc 3′ (SEQ ID NO. 15) and 5′ cccAAGCTTttacagcttacgttttttgcgttta 3′ (SEQ ID NO. 16), respectively. The DNA sequence of the isolated DNA fragment was confirmed by comparing with the sequence from Gene Bank database. In addition, L1 DNA fragments from different strains of HPV-16 can also be cloned from different clinical samples or sources.
[0099]The obtained 1596 base pair (b.p.) DNA fragment was sub-cloned into a baculovirus expression system in order to express a recombinant HPV-16 L1 protein. The D...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com