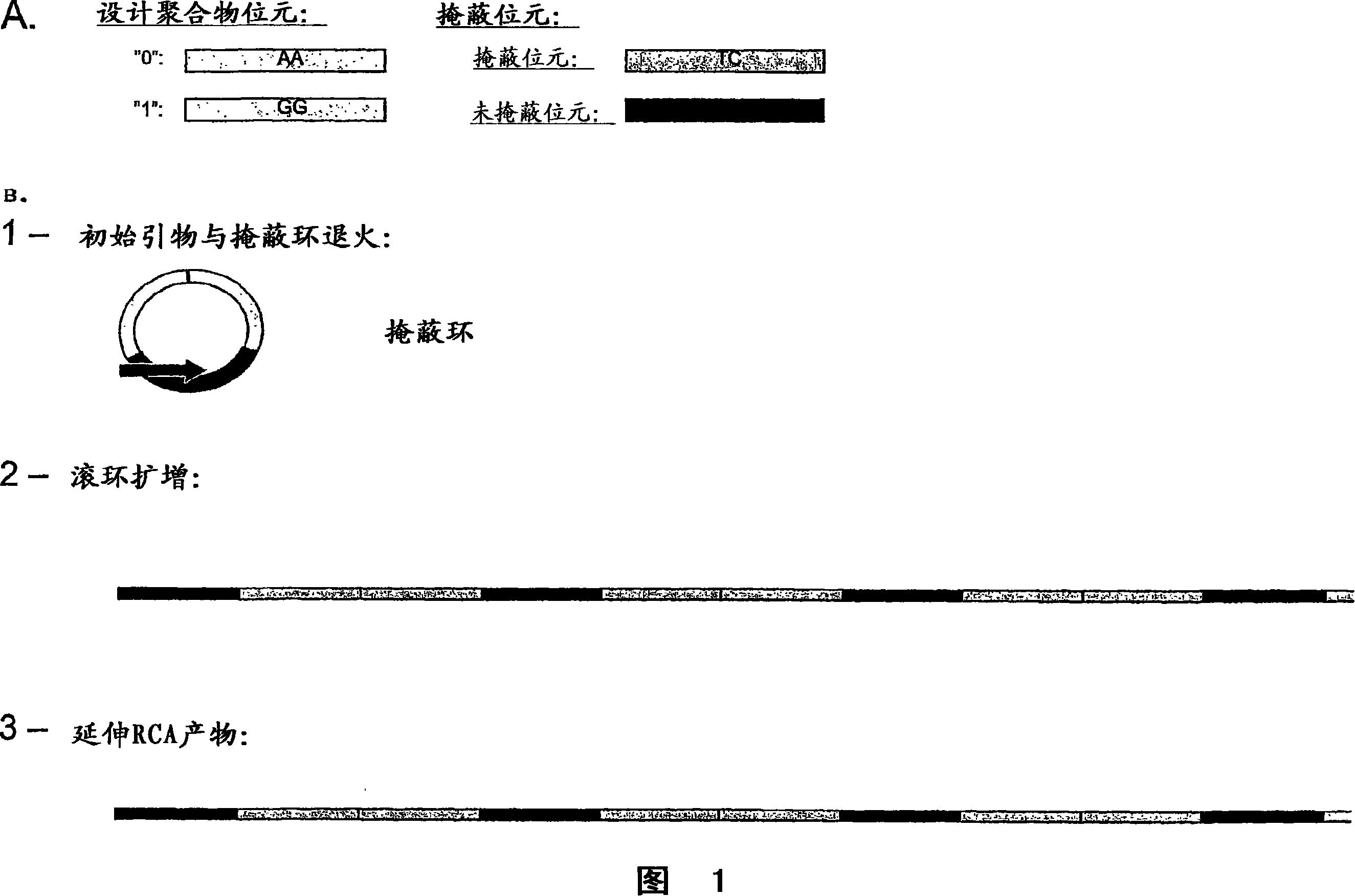
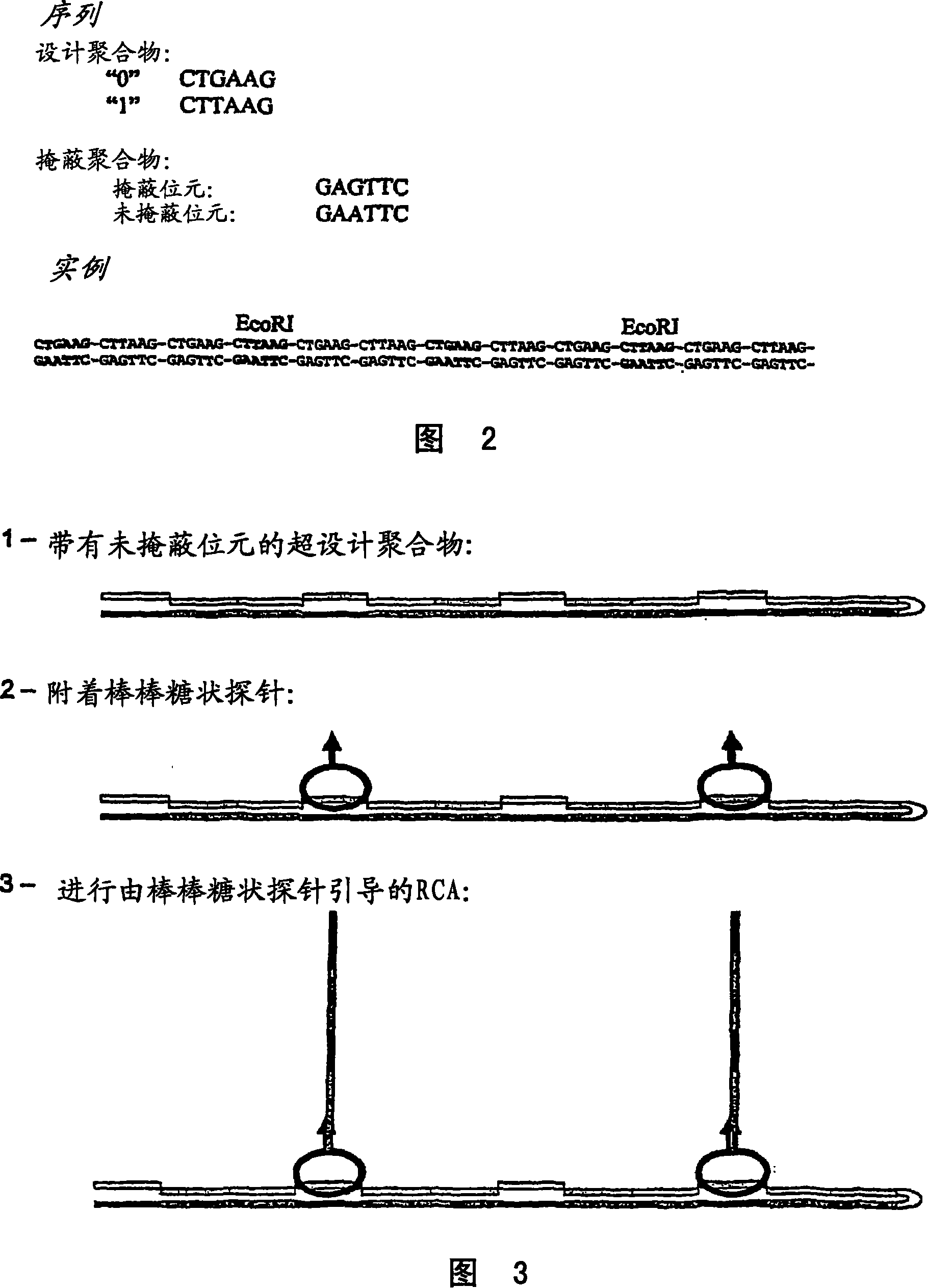
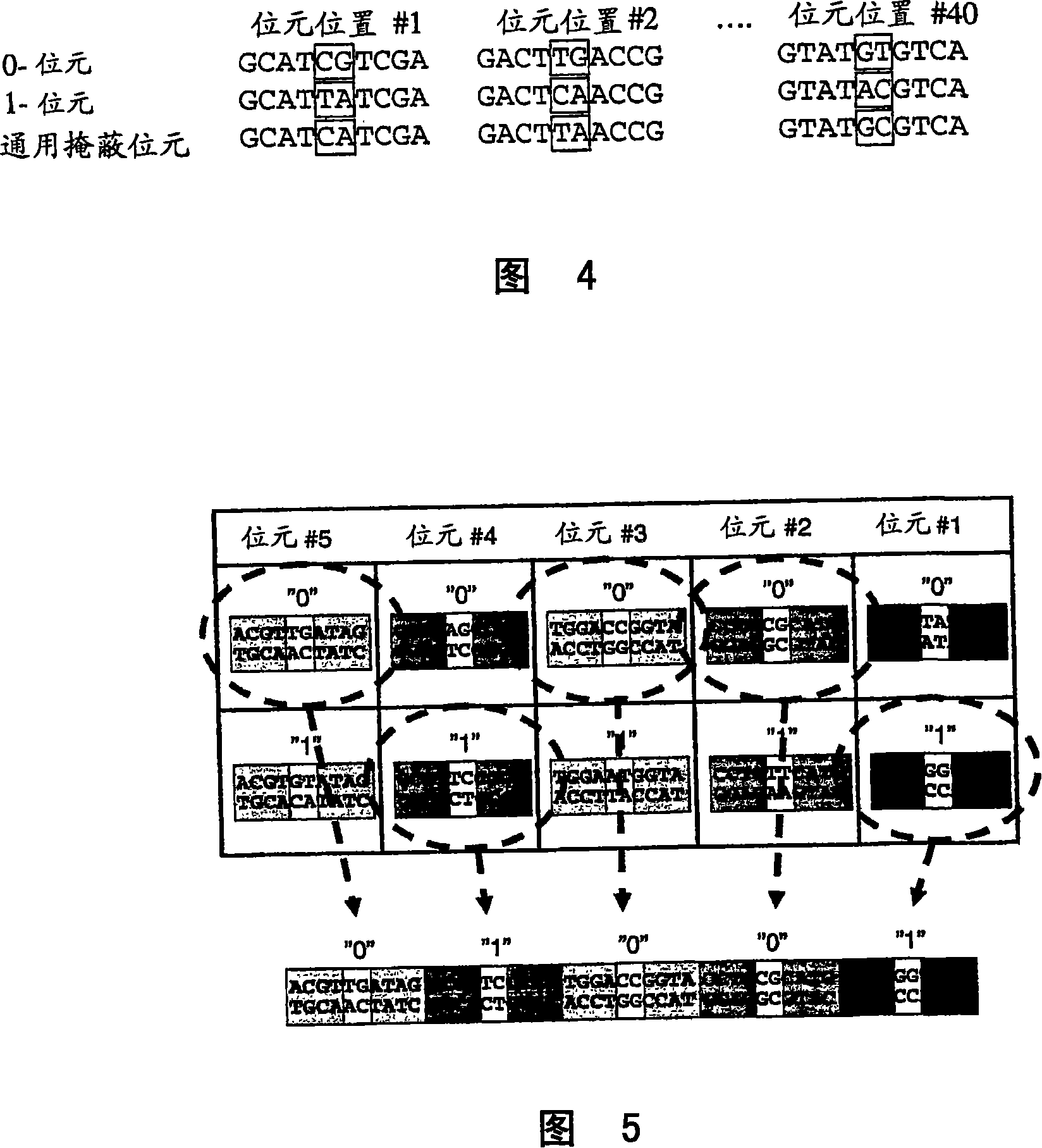
Method for preparing polynucleotides for analysis
A technology of polynucleotides and target polynucleotides, applied in biochemical equipment and methods, measurement/inspection of microorganisms, etc., can solve problems such as readout step obstruction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0086] To illustrate the "rollback" principle, a 114nt single-stranded molecule was used as the second polynucleotide substrate and the 38bp circular target template. The second polynucleotide was "anchored" by immobilizing the substrate molecule with biotin on paramagnetic beads coated with 1 [mu]M streptavidin.
[0087] The target molecule is hybridized to the second polynucleotide and phi29 DNA polymerase is added. The polymerase performs an extension reaction using the target as a template. The extended strand is complementary to the second polynucleotide, and according to the rollback theory, the extended strand will hybridize to the second polynucleotide to form a 114bp double-stranded molecule. Depending on the sequence of the target, the double-stranded molecule will contain recognition sites for certain restriction endonucleases (Figures 10, 11 and 12).
[0088] As shown in FIG. 10 , the target having the unit sequence 0100 forms a recognition site for BamHI at bit ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com