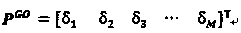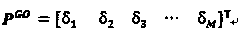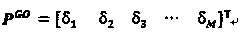A Protein Sequence Representation Method Based on Gene Ontology Information
A protein sequence and gene ontology technology, applied in the field of protein sequence representation based on gene ontology information, can solve the problem of low label positioning prediction rate, and achieve the effect of expanding the scope of use, broad application prospects, and improving the success rate of prediction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0020] In order to make the object, technical solution and advantages of the present invention more clear, the present invention will be further described in detail below in conjunction with the examples. It should be understood that the specific examples described here are only used to explain the present invention, and are not intended to limit the present invention. The example here is an algorithm for predicting subcellular multi-labels of animal proteins.
[0021] Using the new gene ontology information-based protein sequence representation method of the present invention, the specific steps are as follows:
[0022] 1) Use the BLAST program to search the Swiss-Prot database to find all similar protein sequences of protein sequence P.
[0023] Protein P can be directly input to the BLAST tool web page of the Swiss-Prot database, its URL is http: / / www.uniprot.org / blast / , BLAST operation parameters are default, and BLAST can also be downloaded from NCBI for local configurati...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com