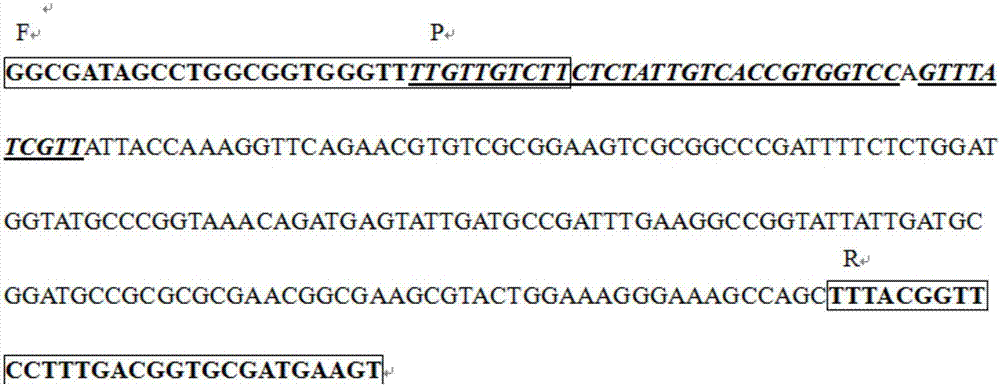Detection method of food-borne pathogenic bacteria salmonella
A technology for food-borne pathogenic bacteria and salmonella, applied in the field of microbial screening, can solve the problems of easy misjudgment, heavy workload, long detection cycle, etc., achieve simple detection, effective salmonellosis outbreak, and inhibit salmonellosis. The effect of the outbreak
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Embodiment 1: Primer and probe design and screening:
[0031] Primers were designed according to the invA gene encoding the Salmonella invasion protein (invasion protein A, invA), and the published invA gene sequence was found through NCBI
[0032] (U43272.1), according to the conserved region, after homology analysis, RPA primers were designed with the gene located at 101-672 as the target fragment.
[0033] Three sets of primer pairs were designed for optimal primer screening. The primer sets and sequences are as follows:
[0034] Group 1:
[0035] Forward primer F1-RPA: 5′-
[0036] TTGTTGTCTTTCTCTATTGTCACCGTGGTCC-3'(30bp)
[0037] Reverse primer R1-RPA: 5′-
[0038] ACTTCATCGCACCGTCAAAGGAACCGTAAA-3′(30bp)
[0039] Product length: 231bp
[0040] Group 2:
[0041] Forward primer F2-RPA: 5′-
[0042] TGTCTTTCTCTATTGTCACCGTGGTCCAGTT-3′(30bp)
[0043] Reverse primer R1-RPA: 5′-
[0044] ACTTCATCGCACCGTCAAAGGAACCGTAAA-3′(30bp)
[0045] Product length: 227bp
...
Embodiment 2
[0076] Detection sensitivity and specificity of embodiment 2 primer probe
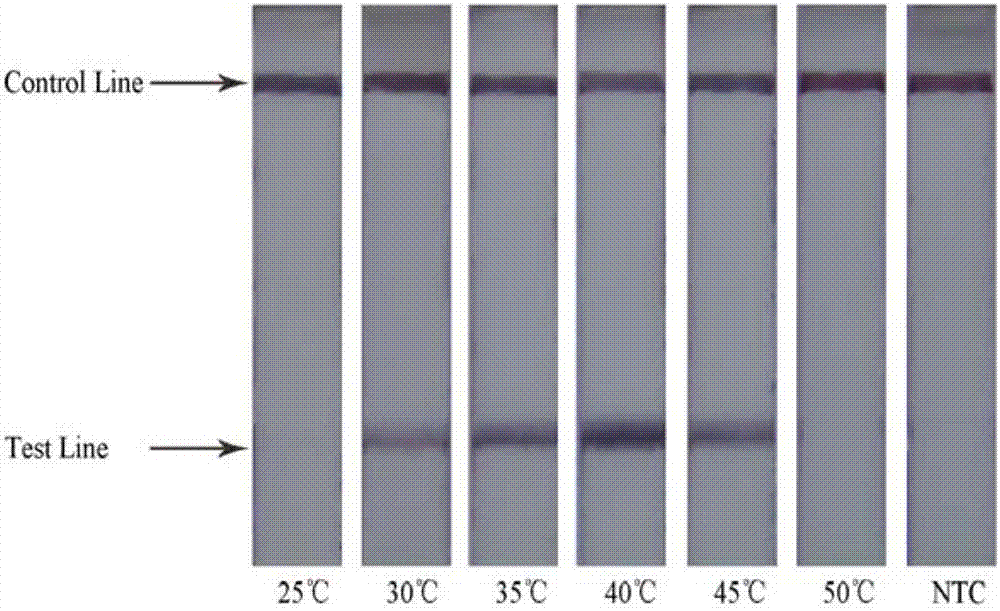
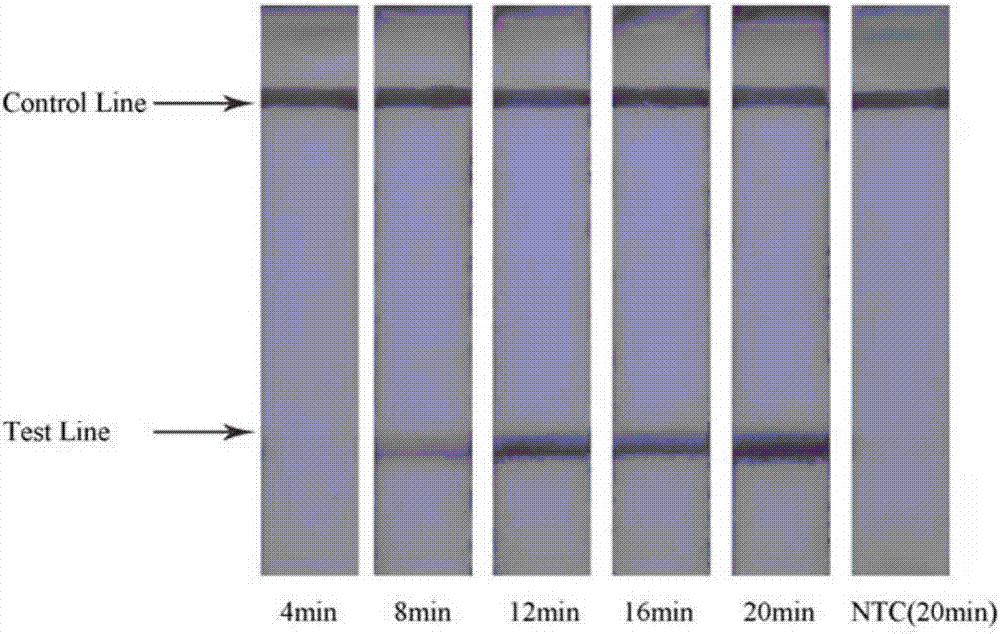
[0077] Set 5 groups of Salmonella DNA templates with different concentrations (DNA concentration gradient is 100, 10-1, 10-2, 10-3, 10-4, 0), and carry out nucleic acid amplification under the optimal conditions of RPA.
[0078] The Salmonella DNA extracted according to the instructions of the DNA extraction kit, the original concentration of the extracted DNA template (511.7ng / μL) was diluted into 10-fold gradients to 100ng / μL, 10ng / μL, 1ng / μL, 100pg / μL, 10pg / μL , 1pg / μL, 100fg / μL, 10fg / μL, 1fg / μL, take 1μL respectively as the reaction template. Carry out nucleic acid amplification according to the aforementioned sample loading method, and detect the LFD of the amplified product:
[0079] The results show that the combination of primers and probes designed in the present invention can ensure the sensitivity of detection, and the detection sensitivity is 5 fg / μL of DNA final concentration, which is eq...
Embodiment 3
[0082] Embodiment 3 to the detection application of actual sample
[0083] 1. Sample purchase:
[0084] Aquatic products come from the aquatic product distribution market, including sea melon seeds (Moerella iridescens), razor clams (Sinonovacula constricta), mussels (Mytilidae), clams (Clam), blood cockles (Sanguinolaria) (10g net content / serving).
[0085] 2. Sample Preparation
[0086] 2.1 Non-frozen samples should be stored in a refrigerator at 7°C to 10°C immediately after collection, and tested as early as possible.
[0087] 2.2 Shellfish take all the contents, including shellfish meat and body fluids. For shellfish, wash the shell in tap water and dry the surface moisture, then open the shell with aseptic operation, and take the corresponding part according to the above requirements.
[0088] 2.3 Take 10 g of the sample by aseptic operation, add 90 mL of Buffered Peptone Water medium (BPW), homogenize with a rotary blade homogenizer at 8 000 r / min for 1 min, or beat ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com