Method for humanbody health assessment anddiseasediagnosis based on human body flora interaction network analysis
A technology of interactive network and flora, applied in the fields of bioinformatics and medical health, can solve problems such as the application limitations of microbial flora research, and achieve the effect of reliable technical support
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0019] Example 1: Comparing the number of special trios in the intestinal flora network of HIV positive and negative persons
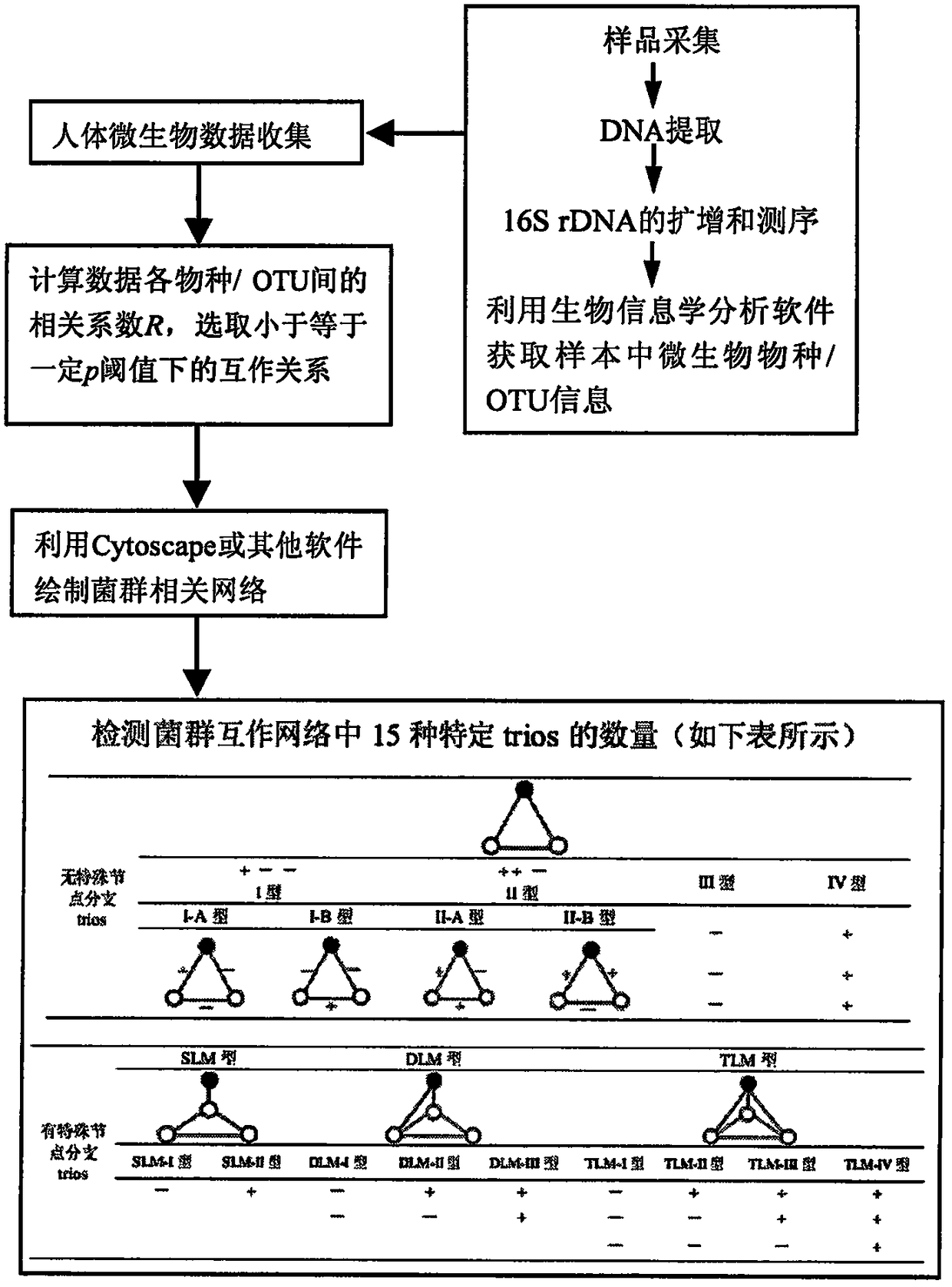
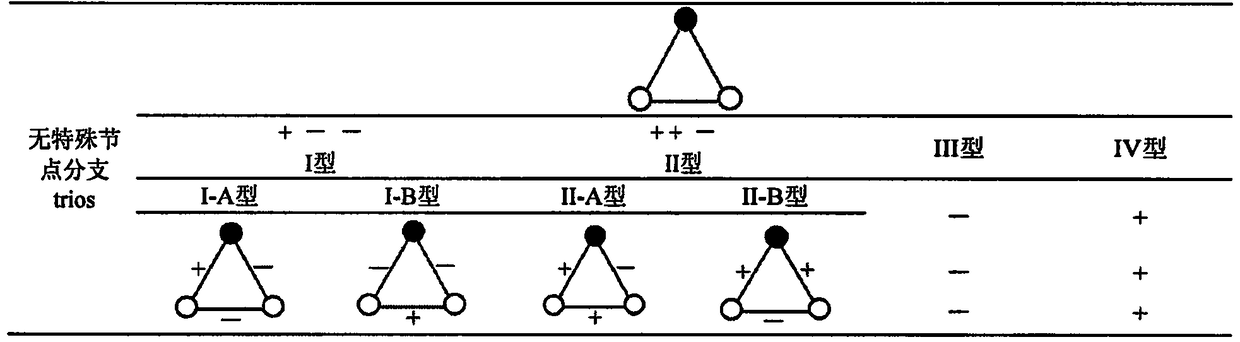
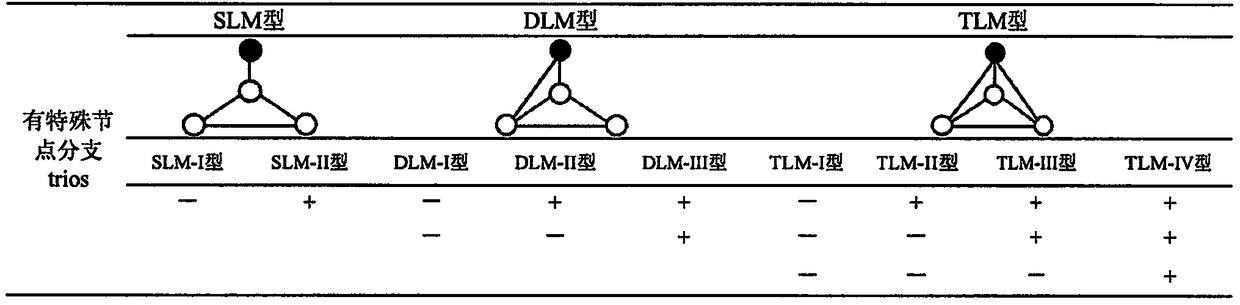
[0020] The calculation process of the present invention is as accompanying drawing figure 1 shown
[0021] Data Sources:
[0022] In 2013, McHardy et al. published the microbial data of HIV-positive and negative individuals. The samples were collected from the intestinal mucosa of 20 HIV patients who had not received antiretroviral treatment and 20 healthy people. 16S rDNA fragments were amplified from the sample DNA using universal primers, sequenced using the Illumina HiSeq 2000 sequencing platform, and 97% similarity of microbial taxonomic units (operational taxonomic unit, OTU) was obtained through subsequent bioinformatics analysis, in which each An OTU represents a species, and the sequencing content of the OTU in each sample represents the species abundance of the OTU in the sample.
[0023] Construction of the interaction network of intestin...
Embodiment 2
[0029] Example 2: Comparing the number of special trios in the oral flora interaction network of smokers and non-smokers
[0030] The calculation process of the present invention is as accompanying drawing figure 1 shown
[0031]Data Sources:
[0032] Lazarevic et al published the oral saliva microbial data of smokers and non-smokers in 2010, and collected oral saliva samples of two non-smoking individuals and three smoking individuals at three time points within 29 days. The 16S rDNA fragment was amplified from the sample DNA using universal primers, and the sequence was sequenced on the Genome Sequencer FLX system, and the microbial taxonomic unit (operational taxonomic unit, OTU) with 97% similarity was obtained through subsequent bioinformatics analysis, in which each An OTU represents a species, and the sequencing content of the OTU in each sample represents the species abundance of the OTU in the sample.
[0033] Construction of oral flora interaction network:
[003...
Embodiment 3
[0039] Example 3: Comparing the number of special trios in the skin flora interaction network of AD patients and healthy people
[0040] The calculation process of the present invention is as accompanying drawing figure 1 shown
[0041] Data Sources:
[0042] The skin microbiome data of AD patients and healthy people published by Kong et al. in 2012, samples were collected from the skin samples of 12 AD patients aged 2-15 years and 11 healthy people. The 16S rDNA fragment was amplified from the sample DNA by using universal primers, and after sequencing, a 97% similarity microbial taxonomic unit (operational taxonomic unit, OTU) was obtained through bioinformatics analysis, wherein each OTU represented a species, and each The sequence content of the OTU in a sample represents the species abundance of the OTU in the sample.
[0043] Construction of skin flora interaction network:
[0044] Based on the species abundance of OTUs, the Spearman correlation coefficient R between...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com