LAMP primer combination for detecting two kinds of cryptococcus and application of LAMP primer combination
A primer combination and primer set technology, applied in the biological field, can solve the problems of high experimental operation requirements and long detection time (about 2.5h-3h).
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0109] Embodiment 1, the preparation of the primer combination that detects two kinds of cryptococci
[0110] The primer set for the detection of two Cryptococci consisted of two LAMP primer sets, each for the detection of one Cryptococcus.
[0111] The primer set for detecting Cryptococcus neoformans is as follows (5'→3'):
[0112] Primer I-F3 (SEQ ID NO: 1): CGAGAAATGTAGGTCCCAATT;
[0113] Primer I-B3 (SEQ ID NO: 2): GCTGCTGATACAAAGATCTAGTG;
[0114] Primer I-FIP (SEQ ID NO: 3): AGCCAAGGCTGGAGCCATTCAAAGTGCTGTCCTCAAATCTACC;
[0115] Primer I-BIP (SEQ ID NO: 4): ATCAGGAAATCTACAACGTCCACGGGAGGCATCGAGCATAGC;
[0116] Primer I-LF (SEQ ID NO: 5): GGACATGAATTCAGTGTAAA;
[0117]Primer I-LB (SEQ ID NO: 6): CTTCTTCTCTGGTCCTTC.
[0118] The primer set for detecting Cryptococcus gattii is as follows (5'→3'):
[0119] Primer I-F3 (SEQ ID NO: 7): AATCTACAACAATAGGCTCTG;
[0120] Primer I-B3 (SEQ ID NO: 8): GGCTGCTGATACAAGGTTC;
[0121] Primer I-FIP (SEQ ID NO: 9): TGGGAAATGGGGATGTAA...
Embodiment 2
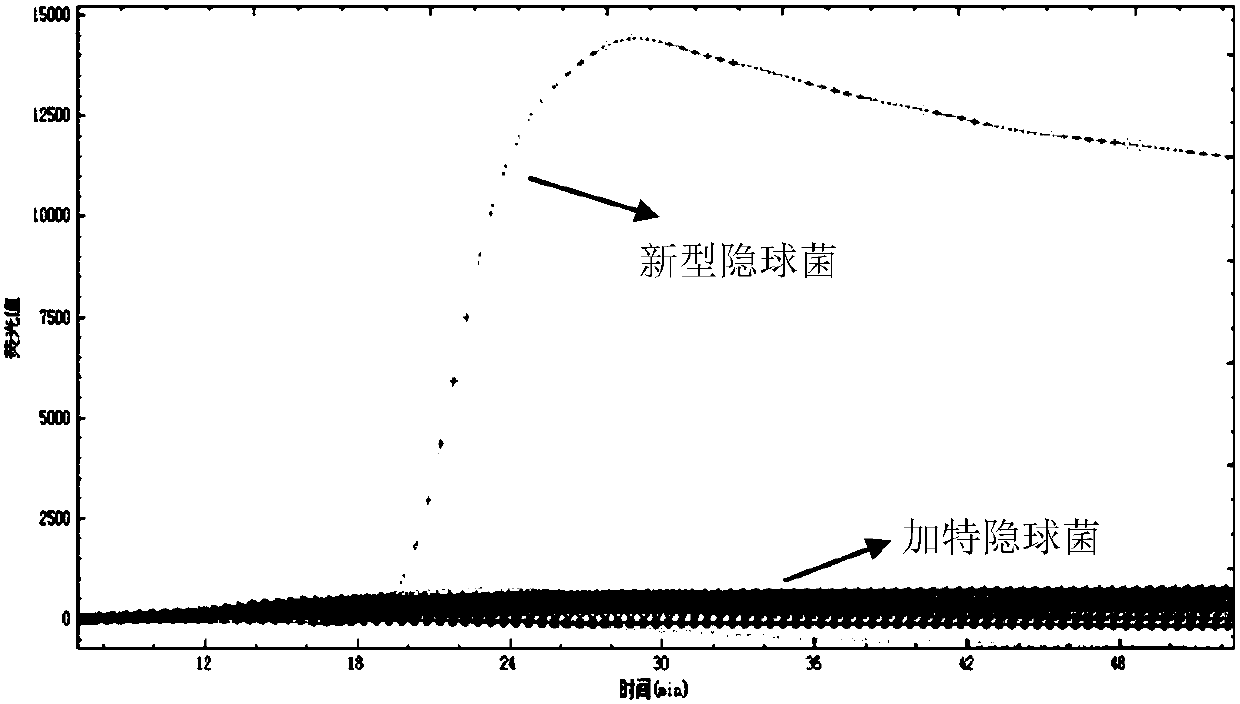
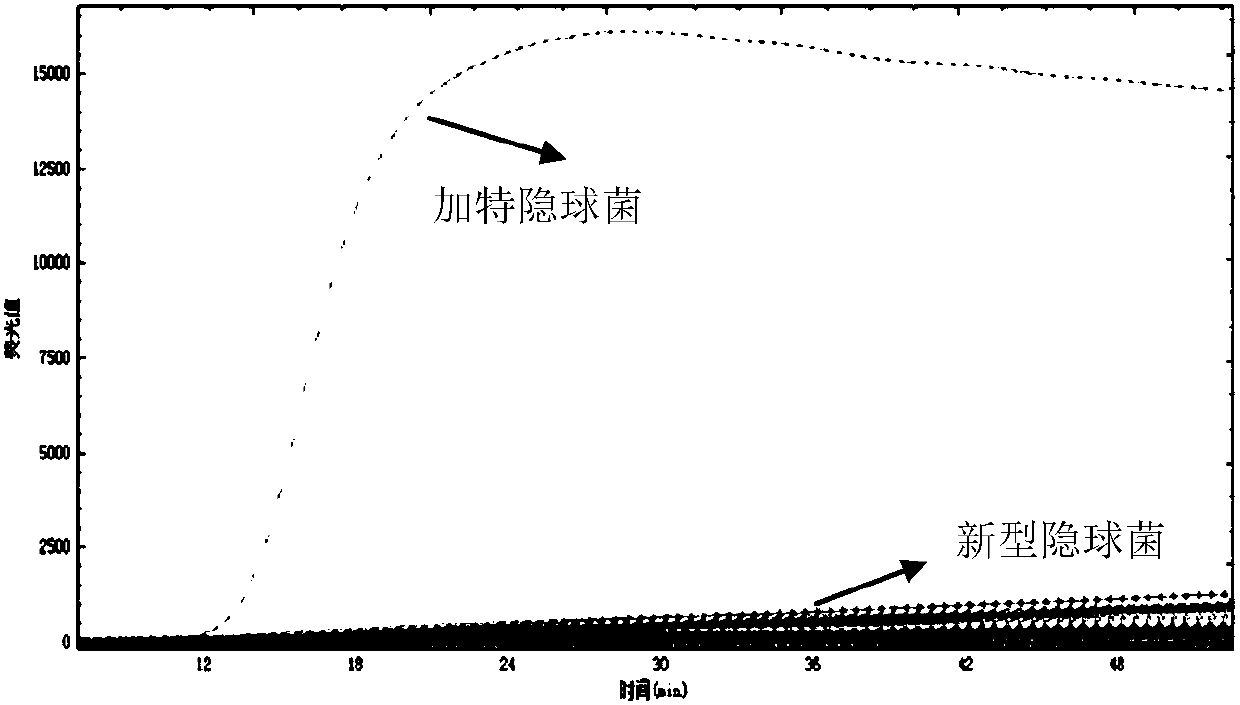
[0130] Embodiment 2, specificity
[0131] 1. Preparation of samples to be tested
[0132] Test sample 1: Genomic DNA of Cryptococcus neoformans (ATCC, strain number: 208821).
[0133] Sample 2 to be tested: Genomic DNA of Cryptococcus gattii (ATCC, strain number: 14248).
[0134] 2. Detection of samples to be tested
[0135] Using the samples to be tested in step 1 as templates, the primer set I and primer set II pairs prepared in Example 1 were used for loop-mediated isothermal amplification detection.
[0136] Reaction system (10 μL): 7.0 μL reaction solution (product of Boao Bio Group Co., Ltd., catalog number CP.440020), 1 μL primer mixture, 1 μL template DNA (5pg-50pg), make up to 10 μL with water. The primer mixture is the mixture composed of each primer in primer group I or primer group II. In the reaction system, the final concentrations of outer primer F3 and outer primer B3 were both 0.5 μM, the final concentrations of inner primer FIP and inner primer BIP were b...
Embodiment 3
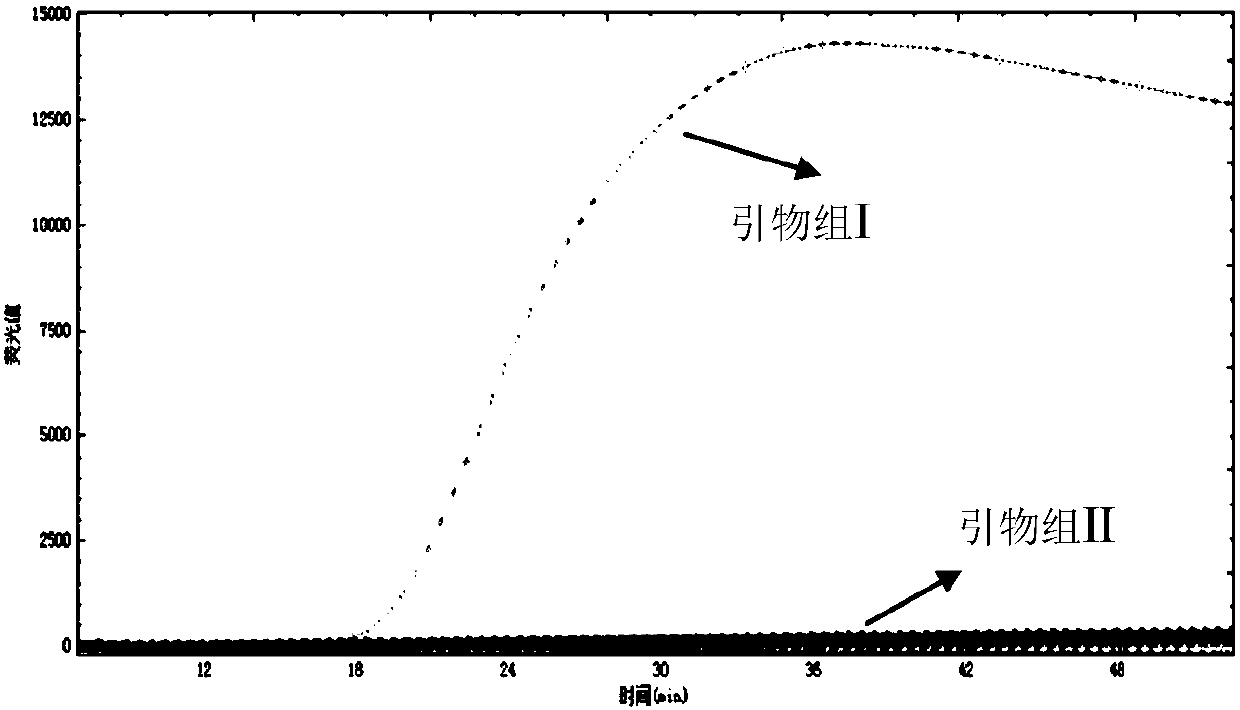
[0143] Embodiment 3, sensitivity
[0144] Sample to be tested 1: Genomic DNA of Cryptococcus neoformans;
[0145] Sample to be tested 2: Genomic DNA of Cryptococcus gattii;
[0146] 1. The sample to be tested is diluted with sterile water to obtain each dilution.
[0147] 2. Using the dilution solution obtained in step 1 as a template, respectively use primer set I and primer set II prepared in Example 1 to perform loop-mediated isothermal amplification.
[0148] When the sample to be tested is the sample to be tested 1, the loop-mediated isothermal amplification is performed using primer set I. When the sample to be tested is the sample to be tested 2, loop-mediated isothermal amplification is performed using primer set II.
[0149] Reaction system (10 μL): 7.0 μL reaction solution (product of Boao Biological Group Co., Ltd., its catalog number is CP.440020), 1 μL primer mixture, 1 μL diluent (1 μL of the genome copy number contained in the diluent is 10 3 , 5×10 2 、10 ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap