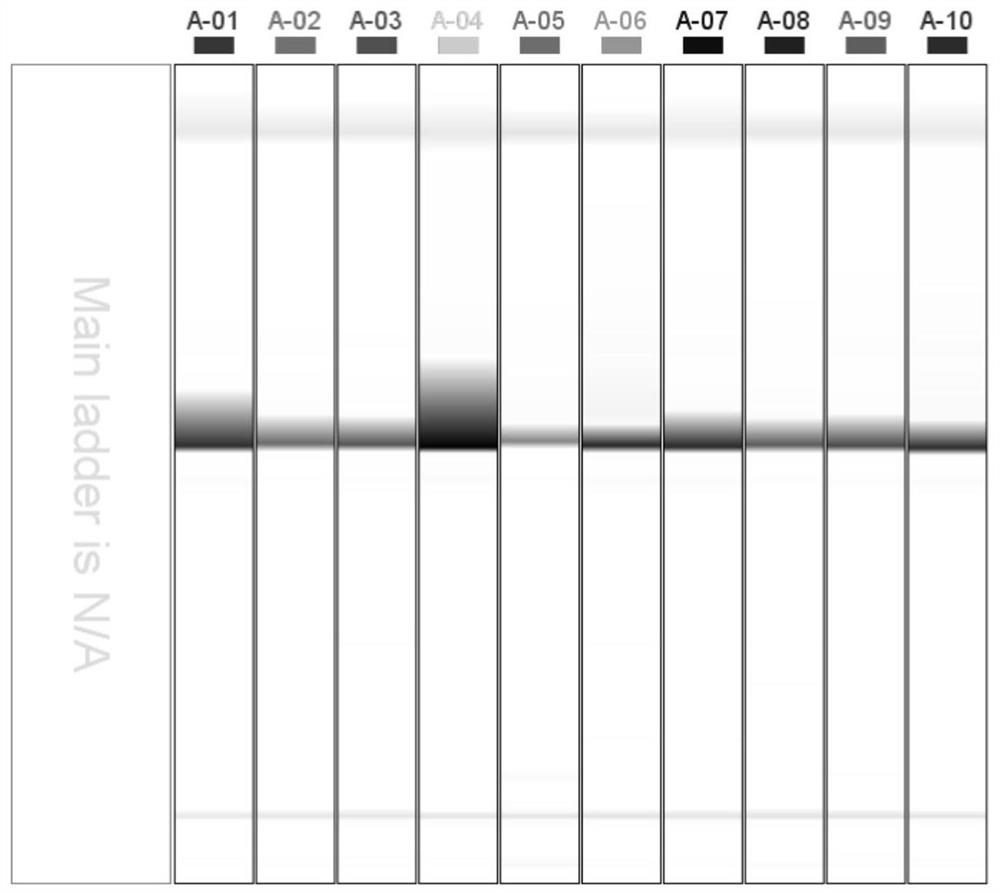
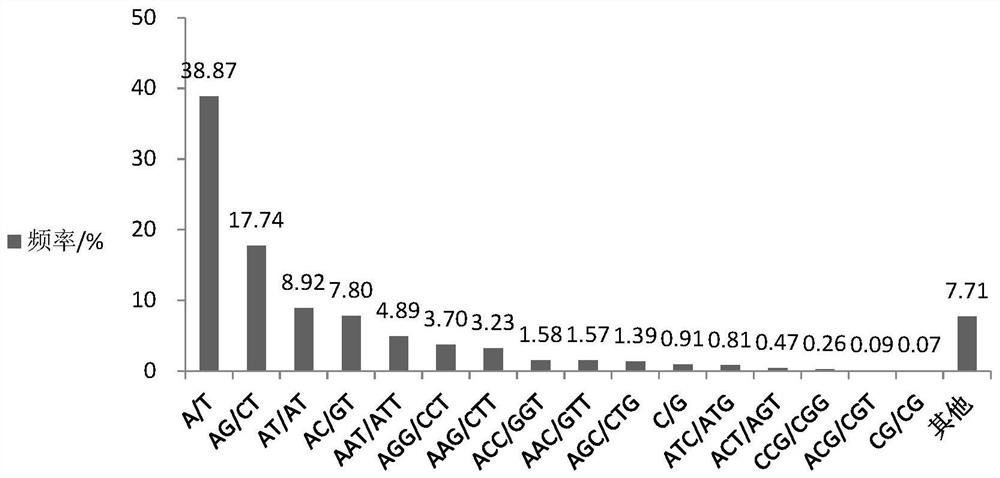
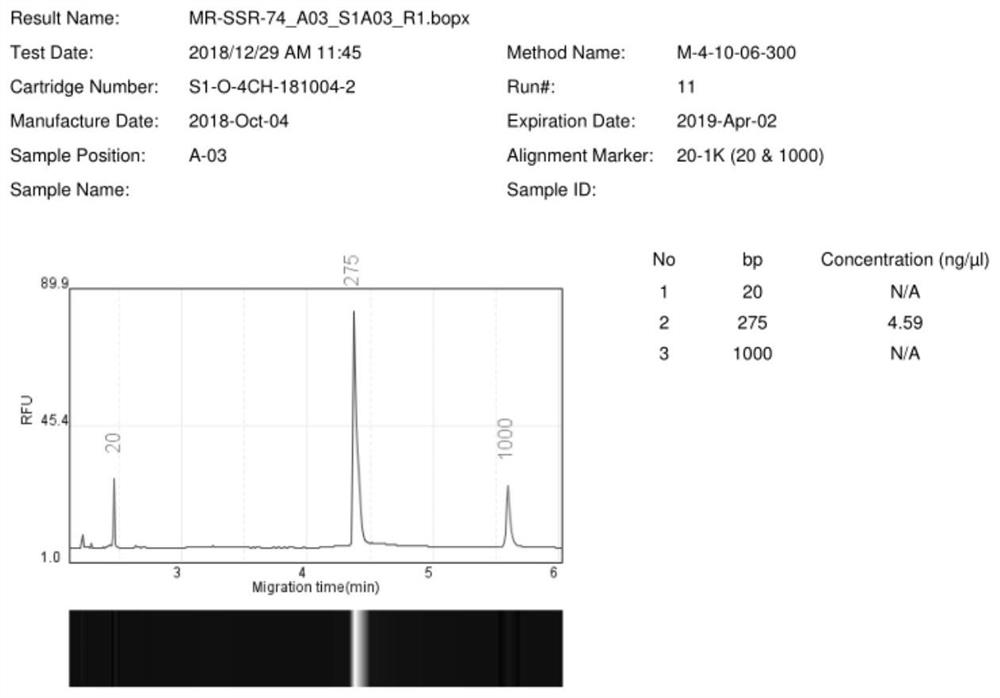
A primer set developed based on the transcriptome sequence of Macrobrachium rosenbergii and its application
A technology of transcriptome sequence and Macrobrachium rosenbergii, applied in the field of SSR primer sets, can solve the problems of limiting the analysis of genetic diversity of Macrobrachium rosenbergii, and achieve the effect of enriching marker sources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Example 1 Transcriptome sequencing
[0034] Randomly selected ovarian tissue from 3 healthy individuals of Macrobrachium rosenbergii, using ReagentInvitrogen extracts RNA. Take equal amounts of RNA and mix them, construct a genomic library and sequence, and entrust Shanghai Lingen Biotechnology Co., Ltd. to complete the sequencing using the Illumina HiSeq 2500 sequencing platform.
[0035] Table 1 Statistics of transcriptome data of Macrobrachium rosenbergii
[0036]
Embodiment 2
[0037] Screening and identification of embodiment 2 SSR sequences
[0038] Use MISA v1.0 (http: / / pgrc.ipk-gatersleben.de / misa) to perform SSR detection on Unigene, and the corresponding minimum number of repeats for each Unit size is the number of repeats when a single nucleotide is used as the repeating unit At least 12 times can be detected; when the repeat unit is dinucleotide, the minimum repeat number is 6; when the repeat unit is trinucleotide, the repeat number is at least 5; when the repeat unit is tetranucleotide, the repeat number is at least 5; When a unit is used, the repeat number is at least 5; when a pentanucleotide is used as a repeat unit, the repeat number is at least 4; when a hexanucleotide is used as a repeat unit, the repeat number is at least 4.
[0039] Finally, statistical analysis was performed on the occurrence frequency of SSR, the type of repeating primitive, the number of repeats and its polymorphism, and the results are shown in Table 2.
[0040...
Embodiment 3
[0045] Design and synthesis of embodiment 3SSR primers
[0046] According to the sequences and sites detected in Example 2, use primer3 (https: / / sourceforge.net / projects / primer3 / ) to design SSR primers in batches. Primer design criteria are: there is no SSR in the primer sequence; the sequence is in the DNA conservative sequence region; the length is between 15-30bp; there is no complementary sequence in the upstream and downstream primers; there is no complementary sequence in the primer itself; the primer annealing temperature (Tm) is 45 Between -60°C; the difference between the Tm values of the upstream and downstream primers is ≤5°C; the GC content is between 40% and 60%; the product size is 100-400bp. Primers for 2-6 nucleotide repeat motifs were synthesized by Shanghai Sangon Bioengineering Co., Ltd. and used for SSR primer screening.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com