A SNP molecular marker, PCR primer and application for identifying Mycobacterium tuberculosis complex
A technology of Mycobacterium tuberculosis and molecular markers, which is applied in the field of SNP molecular markers for identifying Mycobacterium tuberculosis complexes, can solve problems such as inability to distinguish strains, and achieve the effects of not needing a long wait, improving the accuracy of identification, and comprehensively identifying
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Example 1: SNP molecular markers
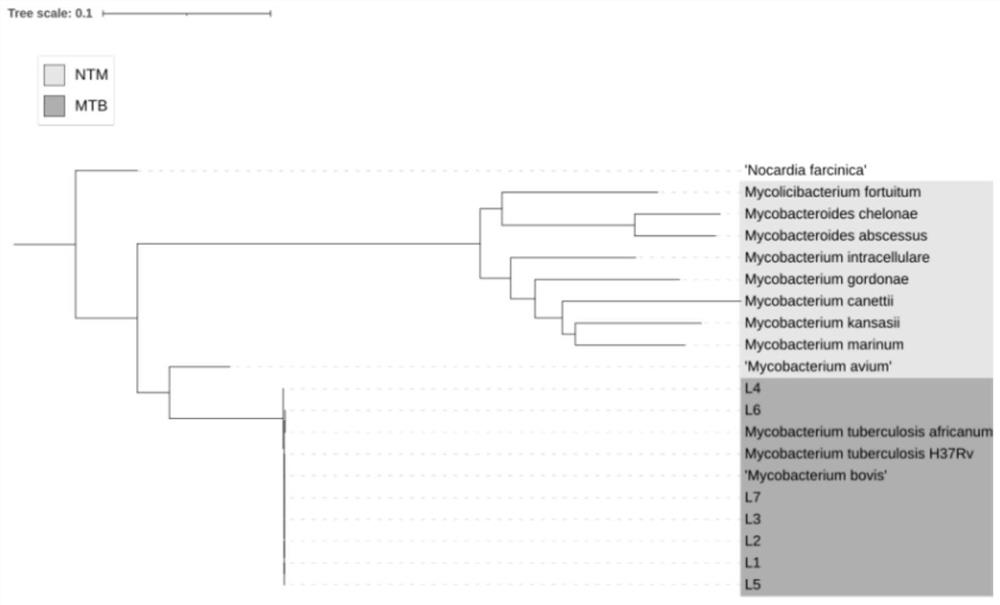
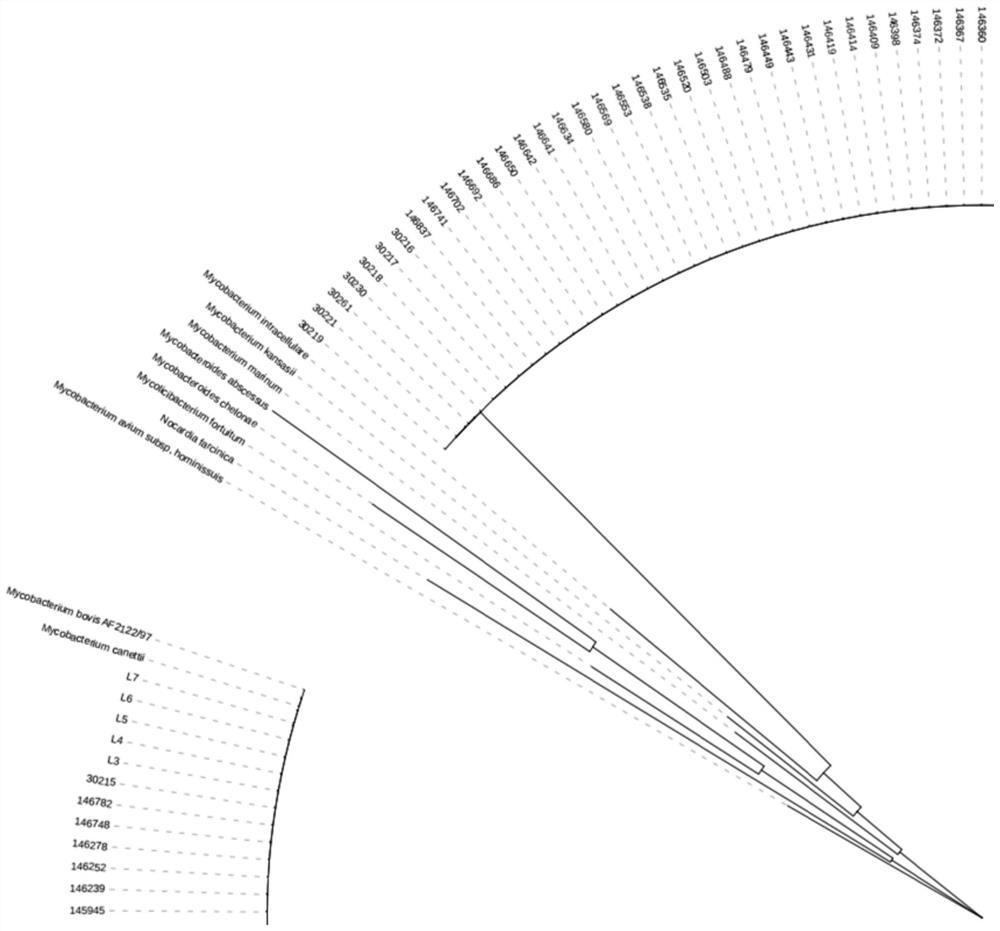
[0062] The SNP molecular markers selected in the present invention are located on the Hsp65 gene and the rpsA gene. The selected genes are highly conserved within Mycobacterium and have strong diversity among different NTMs. The specific method is to divide the target gene into 10 longer fragments by the online analysis software MEME, and follow the internal differences of MTBC as much as possible. In principle, the difference between MTBC and NTM is as large as possible, and the internal fragment of the gene is selected as the alternative amplification region. Import the selected fragment into MEGA7.0 software for gene alignment, and mark the variable site, that is, SNP, following the principle of SNP diversity at the same site as strong as possible (with two or more base types) ) to filter for backup. The phylogenetic tree constructed according to the target gene shows that the difference is more significant, see figure 1 .
[00...
Embodiment 2
[0083] Example 2: PCR detection of the species of mycobacteria to be tested
[0084] Detection method:
[0085] 1) Extract the genomic DNA of the sample to be tested;
[0086] 2) Take the extracted genomic DNA as a template, utilize the primers shown in SEQ ID No.3-4 and the primers shown in SEQ ID No.5-6 respectively, and amplify the DNA fragment containing the core SNP mark by PCR reaction respectively;
[0087] 3) Detecting PCR amplification products, sequencing the amplification products, analyzing the sequencing results, and interpreting the polymorphism at the SNP site.
[0088] Table 2. PCR primer pairs
[0089]
[0090] SEQ ID No. 1:
[0091] GTGATCAGCGAAGAGGTCGGCCTGACGCTGGAGAACGCCGACCTGTCGCTGCTAGGCAAGGCCCGCAAGGTCGTGGTCACCAAGGACGAGACCACCATCGTCGAGGGCGCCGGTGACACCGACGCCATCGCCGGACGAGTGGCCCAGATCCGCCAGGAGATCGAGAACAGCGACTCCGACTACGACCGTGAGAAGCTGCAGGAGCGGCTGGCCAAGCTGGCCGGTGGTGTCGCGGTGATCAAGGCCGGTGCCGCCACCGAGGTCGAACTCAAGGAGCG;
[0092] SEQ ID No. 2:
[0093]ATCCGAAAGGGTG...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com