A fully human anti-new coronavirus neutralizing antibody zw2g10 and its application
A fully human, monoclonal antibody technology, applied in the fields of microbiology and immunology, can solve problems such as loss of neutralizing activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Screening and preparation of human anti-SARS-CoV-2 monoclonal antibodies.
[0037] 1. Collection of blood samples.
[0038] After obtaining informed consent, 20 mL of blood samples were collected 14 days after the second immunization of the recombinant new coronavirus vaccine vaccinators for subsequent experiments.
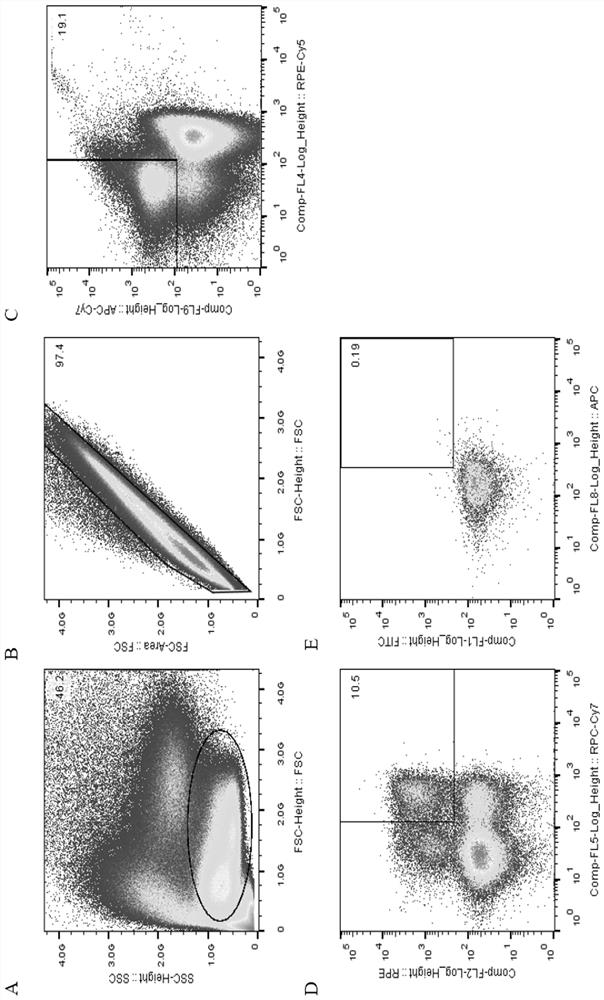
[0039] 2. Sorting memory B cells by flow cytometry.
[0040] The collected blood samples were separated from PBMC by Ficoll density gradient centrifugation, and the process was as follows:
[0041] 1) Take fresh anticoagulated whole blood and anticoagulate with EDTA.
[0042] 2) Add the same volume of separation liquid as the blood sample into the centrifuge tube, spread the blood sample above the liquid surface of the separation liquid, and keep the interface between the two liquid surfaces clear.
[0043] 3) Trim, room temperature, horizontal rotor 800g, acceleration and deceleration speed 3, centrifuge for 30min.
[0044] 4) After centrifug...
Embodiment 2
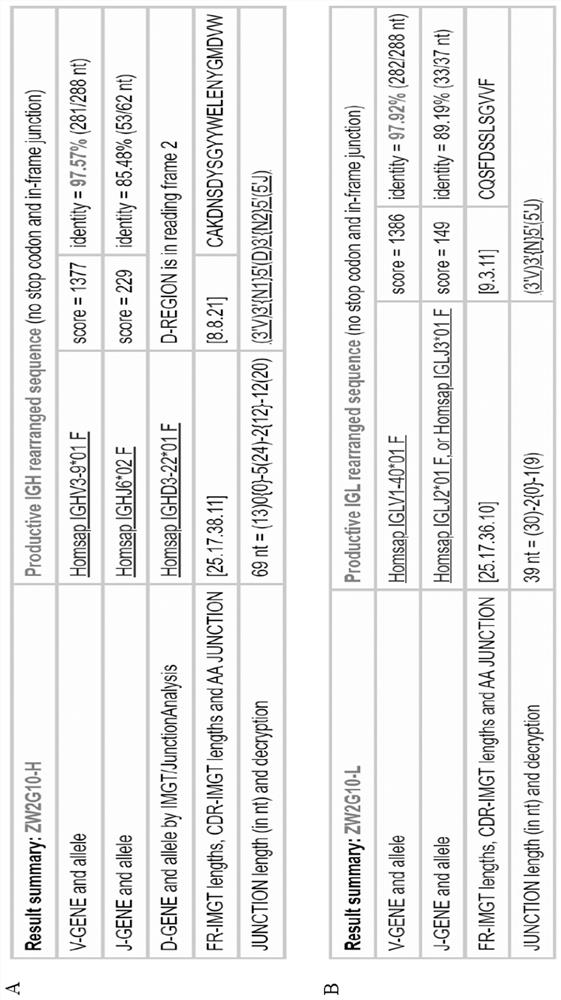
[0118] Example 2. Analysis of the epitope recognized by antibody ZW2G10.
[0119] 1) Coating: Take the recombinant SARS-CoV-2WT S-ECD antigen, S1 antigen, RBD antigen and S2 antigen on the 96-well enzyme-linked plate the day before the experiment and dilute to a concentration of 2 μg / mL with the coating solution, and coat the enzyme label plate, 100 μL per well, and coat overnight at 4°C.
[0120] 2) Blocking: Wash 3 times with a plate washer (BIO-TEK, 405_LS) on the day of the experiment, add 100 μL of blocking solution to each well, and incubate at 37° C. for 1 hour.
[0121]3) Sample incubation: wash the plate 3 times, add 100 μL of diluent to each well except the first well, dilute the antibody to 1 μg / mL in the first well, 4-fold serial dilution, 100 μL / well, set up three replicate wells for each antibody, Incubate at 37°C for 1 h.
[0122] 4) Secondary antibody incubation: wash the plate 3 times, dilute the HPR-labeled goat anti-human IgG secondary antibody (Abcam, ab9...
Embodiment 3
[0128] Example 3: Identification of cross-binding activity of antibody ZW2G10.
[0129] The cross-binding activity between ZW2G10 and SARS-CoV-2 Variants of concern (Variants of concern) S protein was identified, the method was the same as above, and the results were as follows Figure 7 shown. ZW2G10 specifically binds to the S-ECD protein of Alpha strain, Beta strain, Gamma strain, Delta strain, and Omicron strain, and exhibits a dose-response relationship. The results showed that monoclonal antibody ZW2G10 could cross-link the S-ECD protein of Alpha strain, Beta strain, Gamma strain, Delta strain and Omicron strain.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com