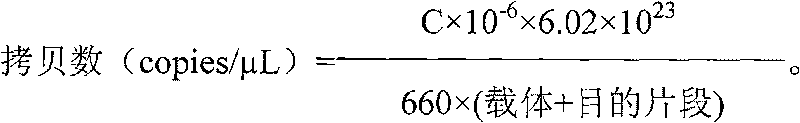Primer for detecting sulfate reduction bacterium
A sulfate and gene sequence technology, applied in the direction of biological testing, material inspection products, etc., can solve the problems of high non-specificity, low detection results, long detection time, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
[0009] Specific embodiment one: The upstream primer gene sequence (FP) for detecting sulfate-reducing bacteria in this embodiment is 5'-GTTCCCTGCTCGTGCCCT-3'; the downstream primer gene sequence (RP) is 5'-TTCCTTGAAGAAGATGTACGGGTT-3'.
[0010] In this embodiment, the detection primers are specifically designed and synthesized according to the conserved structural framework of the DSR gene in sulfate-reducing bacteria.
specific Embodiment approach 2
[0011] Specific embodiment 2: The fluorescent probe used to detect sulfate-reducing bacteria in this embodiment consists of a reporter fluorescent group, a quencher fluorescent group and a probe gene sequence; the probe gene sequence is 5'-AATGGTGGATGGAAGAAGGC-3', The reporter fluorescent group labeled at the 5' end of the probe gene sequence is FAM, and the quencher fluorescent group labeled at the 3' end of the probe gene sequence is TAMRA.
[0012] The gene sequence of the fluorescent probe in this embodiment is specifically designed and synthesized according to the conserved structural framework of the DSR gene in sulfate-reducing bacteria.
specific Embodiment approach 3
[0013] Specific embodiment three: this embodiment adopts fluorescent quantitative PCR method to detect sulfate-reducing bacteria:
[0014] 1. Design and synthesis of detection primers and fluorescent probes
[0015] Design and synthesize specific primers and fluorescent probes according to the conserved structural framework of the DSR gene in sulfate-reducing bacteria. The gene sequence of the upstream primer (FP) is 5'-GTTCCCTGCTCGTGCCCT-3'; the gene sequence of the downstream primer (RP) is 5'-TTCCTTGAAGAAGATGTACGGGTT-3'; the gene sequence of the probe is 5'-AATGGTGGATGGAAGAAGGC-3', the gene sequence of the probe is 5' The reporter fluorophore labeled at the 'end is FAM, and the quencher fluorophore labeled at the 3' end of the probe gene sequence is TAMRA. The synthesis of primers and fluorescent probes was done by a bioengineering company.
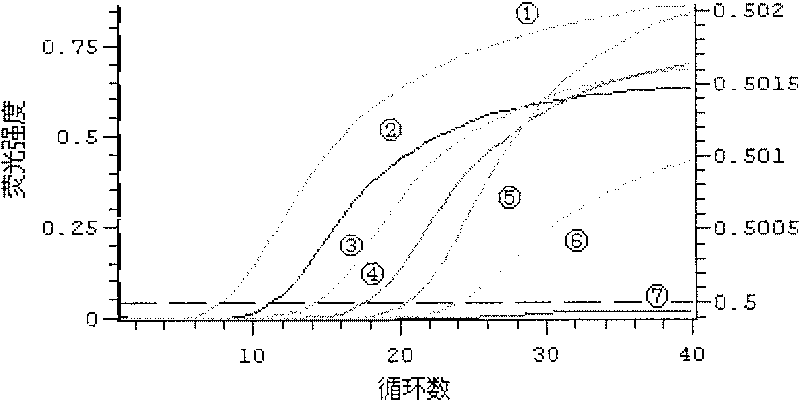
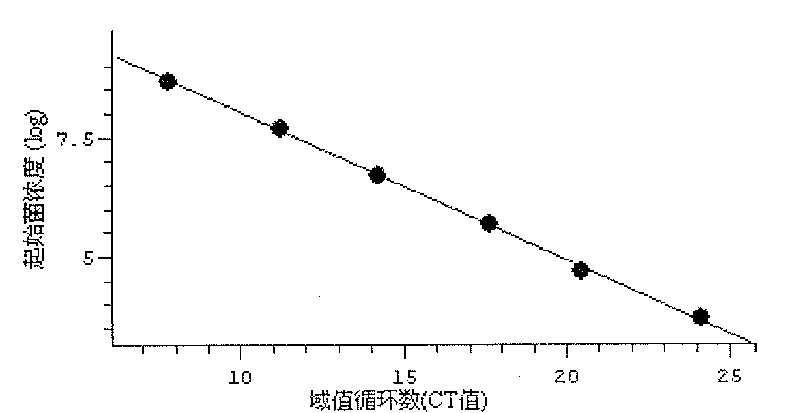
[0016] 2. Primer and fluorescent probe inspection
[0017] (1) Preparation of standard product (sulfate-reducing bacteria) gene fr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com