Soluble fragments of influenza virus pb2 protein capable of binding rna-cap
An influenza virus, soluble technology, applied in the field of soluble fragments of influenza virus PB2 protein that can bind RNA caps, can solve problems such as unknown structure and identity of the binding pocket, controversial results, interference with RNA cap binding, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0164] Example 1: Generation of PB2 expression constructs
[0165] The E. coli codon-optimized PB2 gene (Geneart) SEQ ID NO: 25 based on the influenza A / Victoria / 3 / 1975 (H3N2) PB2 amino acid sequence (dela Luna et al., 1989) was cloned into the pET9a (Novagen) vector , which was modified to introduce a 5'Aatll / AscI and 3'Nsil / NotI restriction site pair for targeted truncating exonuclease III and to provide the 3' sequence encoding the C-terminal biotin acceptor peptide GLNDIFEAQKIEWHE . The first non-directional truncation plasmid library (Tarendeau et al., 2007) was generated from the 3' end, pooled and used as a substrate for the 5' deletion reaction. Linearized plasmids encoding inserts of 150 to 250 amino acids were isolated from agarose gels, religated, and then used to transform E. coli BL21A1 containing RIL plasmids (Stratagene) for use with Alexa488 streptavidin (Invitrogen) Expression screening was performed by hybridization to colony blots (Tarendeau et al., 2007...
Embodiment 2
[0174] Example 2 : Identification of minimal cap-binding fragments
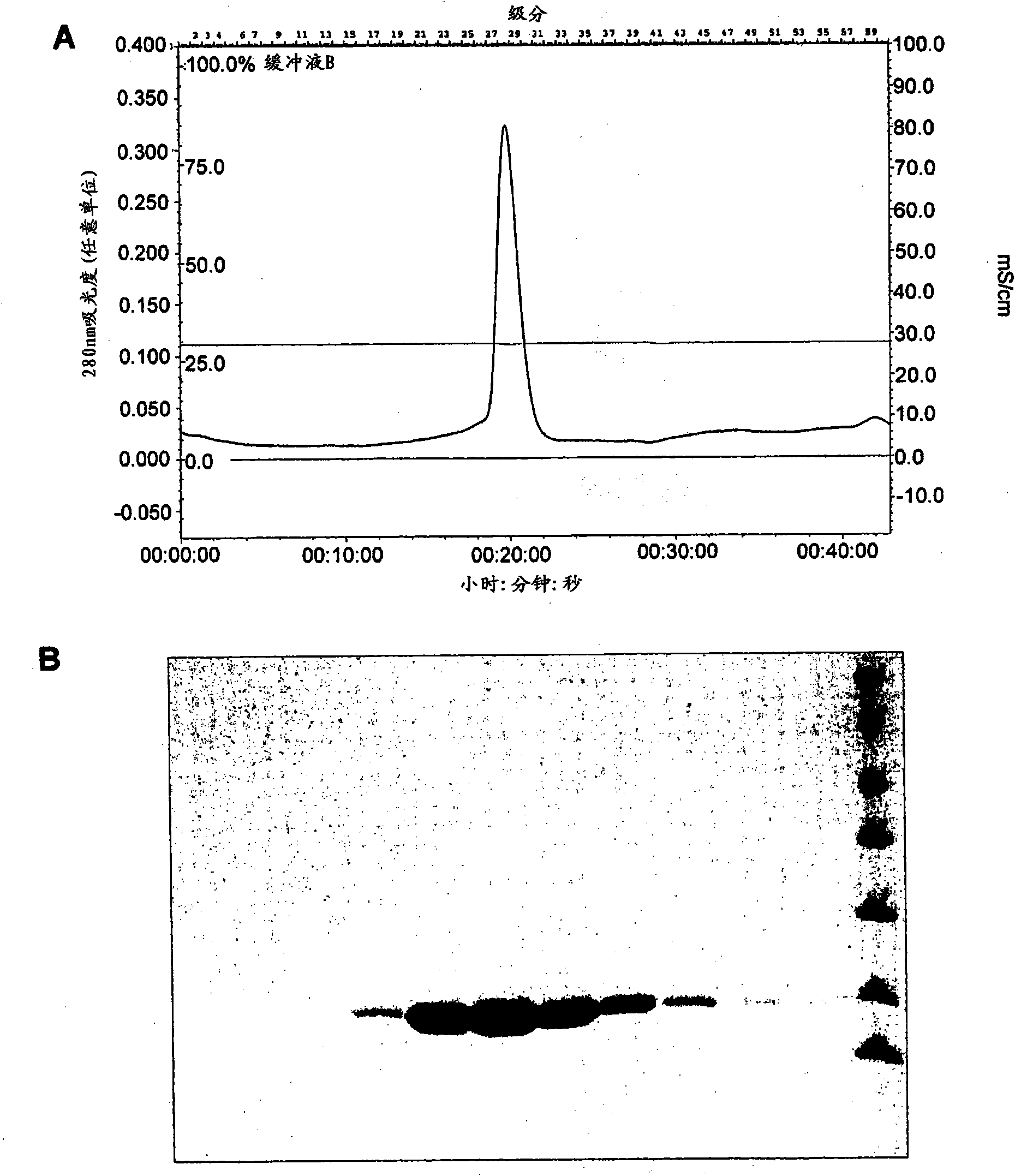
[0175] PB2 polypeptide fragments 235-496, 241-483, 268-483 and 290-483 were expressed in E. coli using the plasmid constructs described in Example 1 and the general expression and purification protocol described in Example 3. Purified PB2 fragment 235-496 was found to be soluble only at low concentrations, while fragments 241-483, 268-483 and 290-483 were more soluble and could specifically bind m 7 GTPsepharose column, indicating cap binding activity. For these assays, 0.2 mg of protein was loaded onto 50 μl of 7-methyl-GTP Sepharose 4B resin (GE Healthcare) and incubated for 3 hours at 4°C for binding. After washing with a buffer containing 50mM Tris.HCl (pH 8.0), 200mM NaCl, 2mM DTT, use a solution containing 1mM m 7 Proteins were eluted by centrifugation in the same buffer as GTP and analyzed on SDS-PAGE. During the course of these experiments, it was noted that smaller degradation fragments can also...
Embodiment 3
[0176] Example 3 : Expression and purification of PB2 fragments
[0177] The plasmids were transformed into E. coli using chemical transformation. The protein was expressed in E. coli strain BL21-CodonPlus-RIL (Stratagene) in LB medium. After induction with 0.2 mM isopropyl-β-thiogalactopyranoside (IPTG) for 16 to 20 hours at 25°C, cells were harvested and resuspended in lysis buffer (50 mM Tris HCl (pH 8.0), 300 mM NaCl , 5mM 2-mercaptoethanol) and sonicated. After centrifugation, the clarified lysate was directly loaded onto a nickel affinity column (chelated sepharose from GE Healthcare loaded with Ni 2+ ion). Wash the Ni-sepharose resin thoroughly with 50mM Tris·HCl (pH 8.0), 1M NaCl, 15mM imidazole, mM 2-mercaptoethanol buffer, and then rinse with 50mM Tris·HCl(pH 8.0), 200mM NaCl, 50mM imidazole, 5mM 2-mercapto Wash with ethanol buffer. Then, the protein was eluted with 50 mM Tris·HCl (pH 8.0), 200 mM NaCl, 0.5 M imidazole, 5 mM 2-mercaptoethanol buffer. Purified...
PUM
| Property | Measurement | Unit |
|---|---|---|
| unit cell dimension | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com