siRNA interference efficiency prediction system based on arm microprocessor
A microprocessor-based, efficient technology, applied in the field of bioinformatics research, can solve problems that do not consider mRNAmotif features, structural features, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
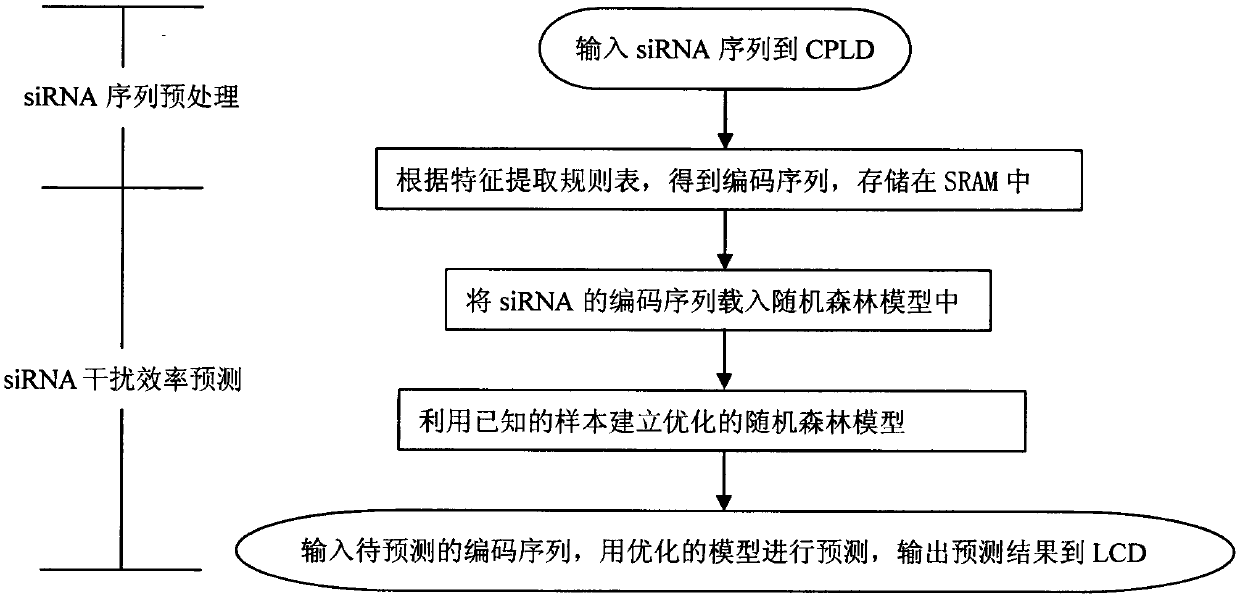
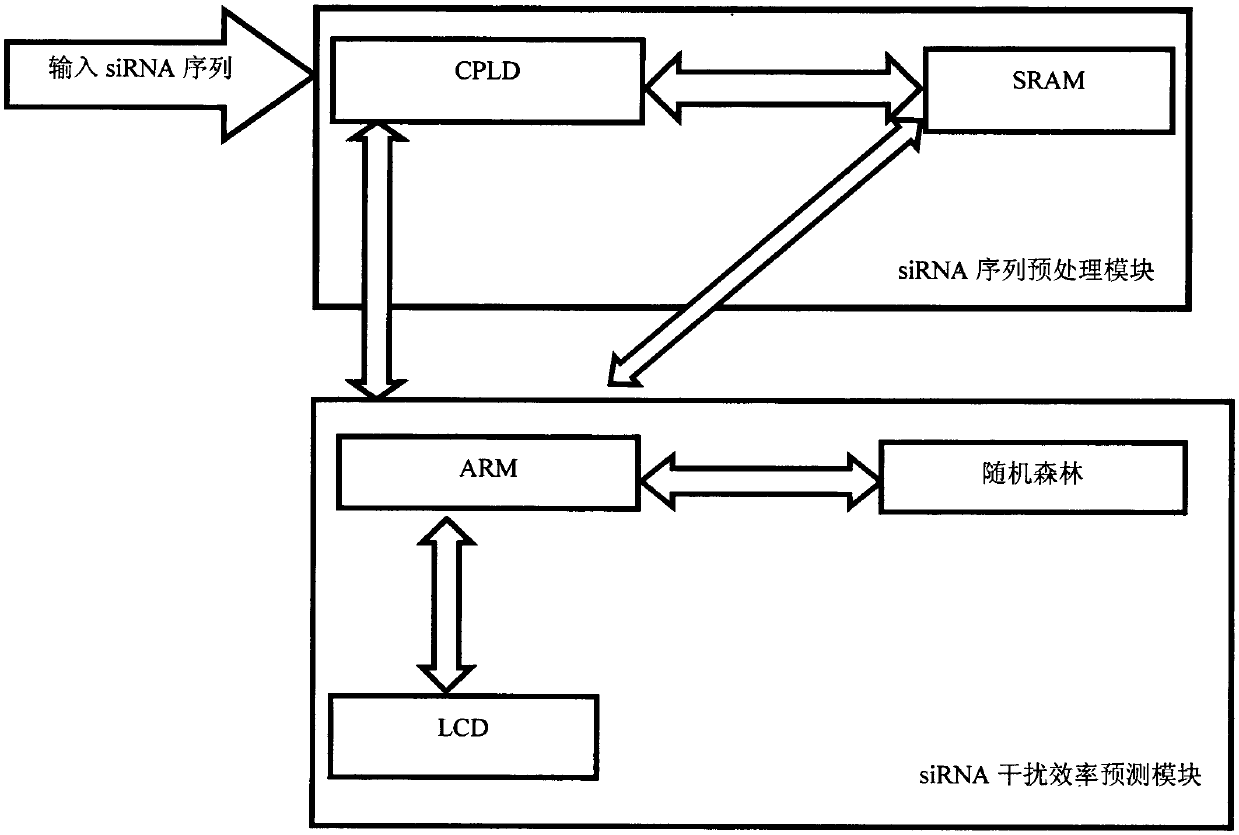
[0041] The present invention is a new method for predicting siRNA interference efficiency based on ARM microprocessor, such as figure 1 As shown, the obtained siRNA sequence is input into CPLD, and the siRNA sequence is encoded through the feature extraction rule table, so that the siRNA sequence is stored in the SRAM in the form of a coding sequence, and then the ARM microprocessor is used to predict the efficiency of siRNA interference. It is necessary to establish a regression model based on random forest first, and adjust the parameters to minimize the generalization error of the model, and then load the digital information corresponding to the siRNA sequence to be predicted into the established random forest model to predict the interference efficiency , and finally output the predicted results to the LCD liquid crystal display.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com