Lipase mutant derived from Talaromyces thermophilic and its application
A lipase and mutant technology, which is applied to a lipase mutant derived from Talaromyces thermophilus and its application field to achieve the effect of improving the activity of the parent
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
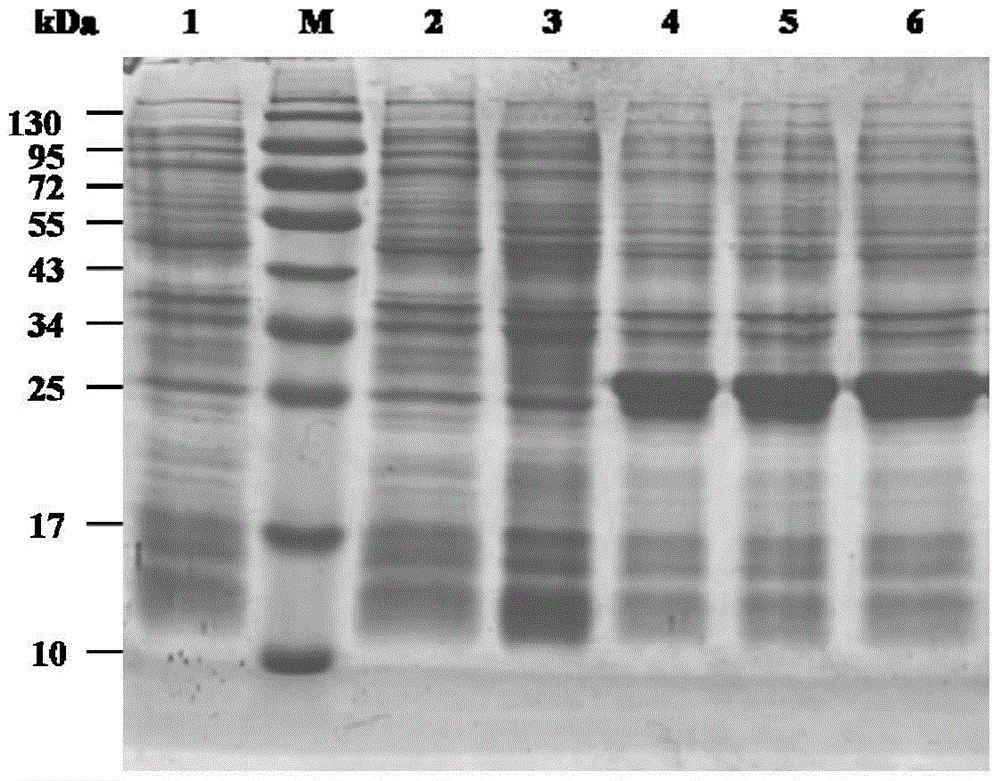
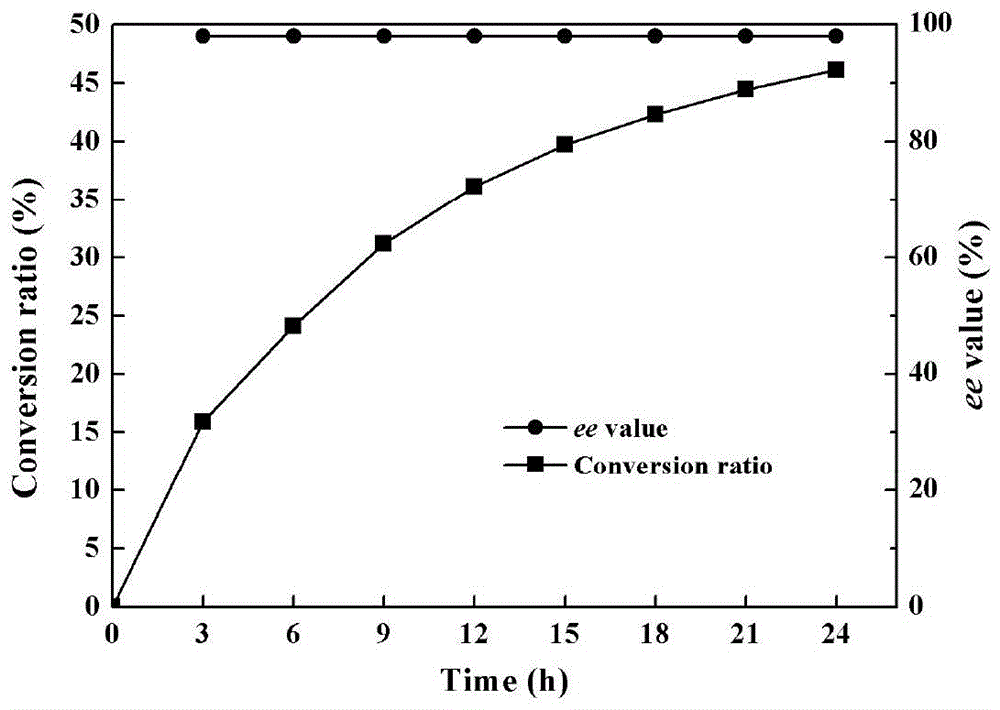
Image
Examples
Embodiment 1
[0023] Example 1: Amplification of parental genes and construction of expression vectors
[0024]The lipase gene was obtained from Talaromyces thermophilus ATCC 20186 (purchased from American Type Culture Collection, American Type Culture Collection) by RT-PCR. According to the Talaromyce thermophilus CTM10.103 lipase gene sequence (GenBank accession no. JF414585) reported in the literature, primers TTL-F and TTL-R (see Table 1) were designed, and the lipase gene was isolated from T.thermophilus ATCC20186 by RT-PCR . Total RNA of T.thermophilus ATCC 20186 was extracted by TRIzol method, using GoScript from Promega TM The reverse transcription kit is used to prepare the first strand of cDNA, and the reaction system and conditions refer to the instructions of the kit. Using the first strand of cDNA as a template, under the action of primers TTL-F and TTL-R, the lipase cDNA sequence was amplified by PCR. The amount of each component in the PCR reaction system (total volume 100...
Embodiment 2
[0025] Example 2: Site-directed saturation mutation of lipase position 83
[0026] Description of site-directed saturation mutagenesis technology reference (Current Protocols in Protein Science26.6.1-26.6.10, 2011; Appl.Microbiol.Biotechnol.2014,98:2473-2483), high-throughput screening technology reference for positive mutants (Appl. Microbiol.Biotechnol.2014,98:2473-2483). The specific process is as follows:
[0027] In order to saturate the Ser at the 83rd position in the parental amino acid sequence, design mutation primers S83-F and S83-R (see Table 1), use the plasmid pET28-TTL constructed in Example 1 as a template, and carry out full-plasmid PCR . The PCR amplification system is: 5×PS buffer 10 μL, dNTP (2.5mM each) 4 μL, mutant primers S83-F and S83-R 0.5 μL each, plasmid pET28-TTL 0.5 μL, PrimeSTAR DNA polymerase 0.5 μL, water up to 50 μL . PCR conditions were pre-denaturation at 98°C for 2 minutes, 25 cycles: 98°C for 10s, 65°C for 10s, 72°C for 6min, and finally...
Embodiment 3
[0028] Example 3: Site-directed saturation mutation of lipase mutant S83T site 58
[0029] Description of site-directed saturation mutagenesis technology reference (Current Protocols in Protein Science26.6.1-26.6.10, 2011; Appl.Microbiol.Biotechnol.2014,98:2473-2483), high-throughput screening technology reference for positive mutants (Appl. Microbiol.Biotechnol.2014,98:2473-2483). The specific process is as follows:
[0030] On the basis of the lipase mutant S83T, the Ser at position 58 was saturatingly mutated, and the mutant primers S58-F and S58-R (see Table 1) were designed, and the plasmid pET28-S83T was used as a template (see Example 2). Whole plasmid amplification. The PCR system is: 10 μL of 5×PS buffer, 4 μL of dNTP (2.5 mM each), 0.5 μL of mutant primers S58-F and S58-R, 0.5 μL of plasmid pET28-S83T, 0.5 μL of PrimeSTAR DNA polymerase, and water to 50 μL. PCR conditions were pre-denaturation at 98°C for 2 minutes, 25 cycles: 98°C for 10s, 65°C for 10s, 72°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com