A kind of pantophyte amidase mutant, gene, engineering bacteria and application thereof
A bacterial amidase and mutant technology, applied in genetic engineering, application, plant genetic improvement and other directions, can solve problems such as low catalytic efficiency, achieve high catalytic efficiency, reduce product inhibition effect, and facilitate extraction and purification.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Site-directed saturation mutation and screening of amidase
[0028] Description of site-directed saturation mutation technology reference (Appl.Microbiol.Biotechnol., 2014,98,2473-2483), reference for high-throughput screening model of positive mutants (CN100370034; Appl.Microbiol.Biotechnol., 2007,74,256-262) description of. The specific process is as follows:
[0029] In the amino acid sequence of Pantoea amidase Pa-Ami (the amino acid sequence is shown in SEQ ID No.2, and the nucleotide sequence is shown in SEQ ID No.1) derived from Pantoea sp. (GenBank No.WP008109374) Glycine (Gly, G) at position 175, threonine at position 301 (Thr, T), alanine at position 305 (Ala, A) and serine at position 309 (Ser, S) were respectively For saturation mutation, design primers for each mutation (see Table 1), and use the plasmid pET28-Pa-Ami cloned with the gene encoding Pantoea amidase Pa-Ami as a template to amplify the whole plasmid. PCR system: 2×phanta Max buffer...
Embodiment 2
[0033] Example 2: Multi-site stacking saturation mutation of amidase mutant G175A
[0034] The saturation mutation technique and the high-throughput screening method for positive mutants are the same as in Example 1. The specific process is as follows:
[0035] On the basis of the amidase mutant G175A, sites including the 301th threonine (Thr, T), the 305th alanine (Ala, A) and the 309th serine (Ser, S) were determined For saturation mutation, the plasmid pET28-G175A of the amidase mutant G175A was used as a template for full plasmid amplification. The PCR system is: 2×phanta Max buffer 25 μL, dNTP mixture (10 mM) 0.75 μL, mutant upstream primer (50 μM) and downstream primer (50 μM) (see Table 1 for primer sequence) each 1 μL, plasmid pET28-G175A 0.5 μL, Phanta Max DNA Polymerase 0.5 μL, ddH 2 O to make up to 50 μL. PCR conditions were pre-denaturation at 95°C for 2 min; denaturation at 95°C for 15 s, annealing at 55-65°C for 15 s, extension at 72°C for 6.5 min, and 30 cyc...
Embodiment 3
[0036] Example 3: Induced expression and purification of parent amidase and amidase mutant engineered bacteria
[0037] (1) Induced expression
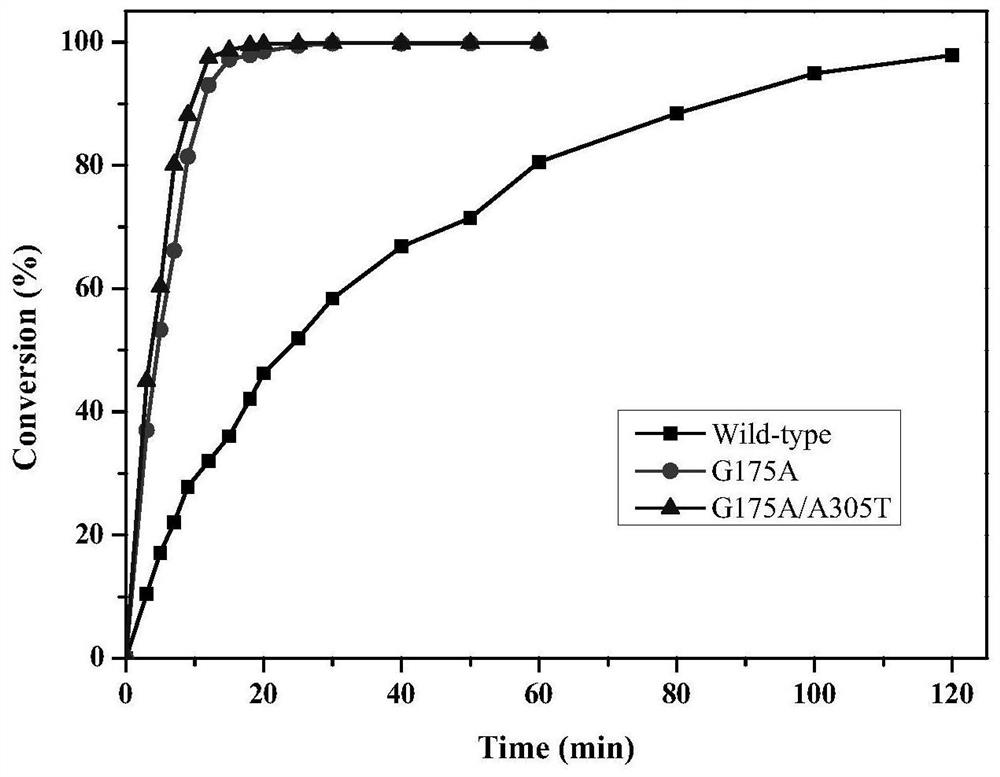
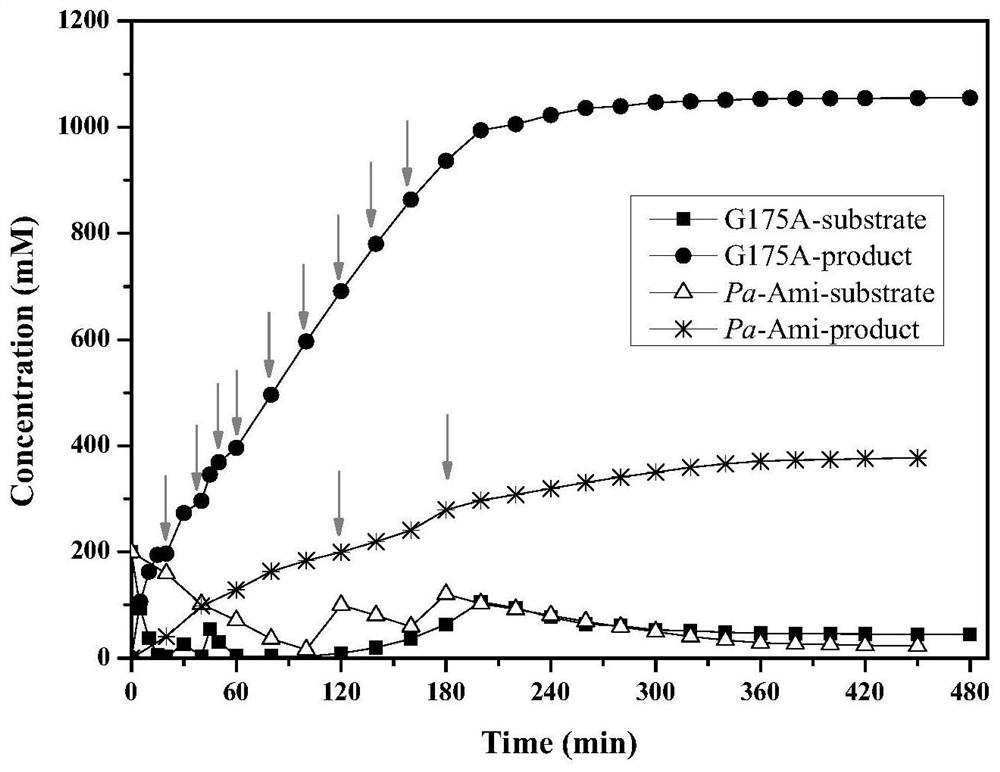
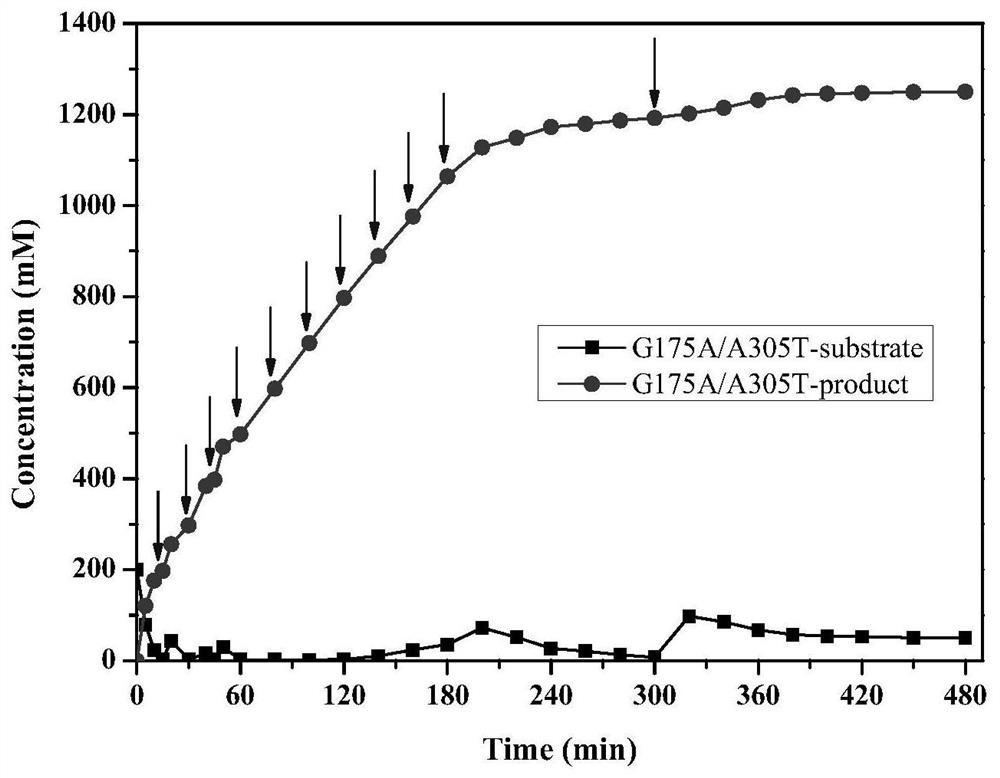
[0038] Will include starting strain E.coli BL21(DE3) / pET28-Pa-Ami and mutant strain E.coli BL21(DE3) / pET28-G175A (Example 1), E.coli BL21(DE3) / pET28-G175A / A305T (Example 2) each positive mutant bacterial strain is inoculated respectively in the LB liquid culture medium containing 50mg / L kanamycin, cultivates 12h under 37 ℃, 150r / min, then with 1% (v / v) Transfer the inoculum to fresh LB liquid medium containing 50mg / L kanamycin, and cultivate at 37°C and 150r / min until the cell concentration OD 600 0.4 to 0.8, then add IPTG with a final concentration of 0.1mM to the medium, induce culture at 28°C and 150r / min for 12h, take the culture and centrifuge at 8000rpm for 15min at 4°C, and collect the wet cells, which can be used for enzyme activity determination and Biocatalytic preparation of 2-chloronicotinic acid.
[0039] (2) Protein p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com