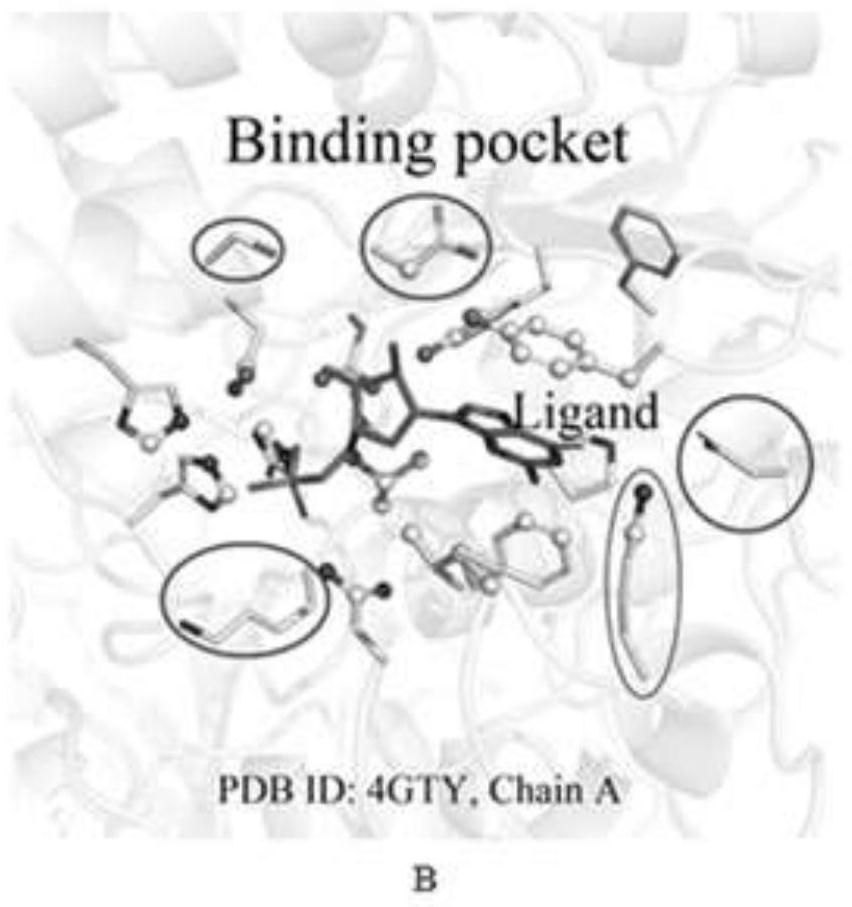
Method for identifying key flexible amino acids on protein small molecule binding pockets
A flexible amino acid and recognition method technology, which is applied in the field of recognition of key flexible amino acids on protein small molecule binding pockets, can solve many problems such as
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0073] In order to better explain the present invention and facilitate understanding, the present invention will be described in detail below through specific embodiments in conjunction with the accompanying drawings.
[0074] In order to better understand the above technical solutions, the following will describe exemplary embodiments of the present invention in more detail with reference to the accompanying drawings. Although exemplary embodiments of the present invention are shown in the drawings, it should be understood that the invention may be embodied in various forms and should not be limited to the embodiments set forth herein. Rather, these embodiments are provided so that the present invention can be more clearly and thoroughly understood, and the scope of the present invention can be fully conveyed to those skilled in the art.
[0075] Specific embodiment description part:
[0076] Based on the above principles, the present invention will (1) classify the protein ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com