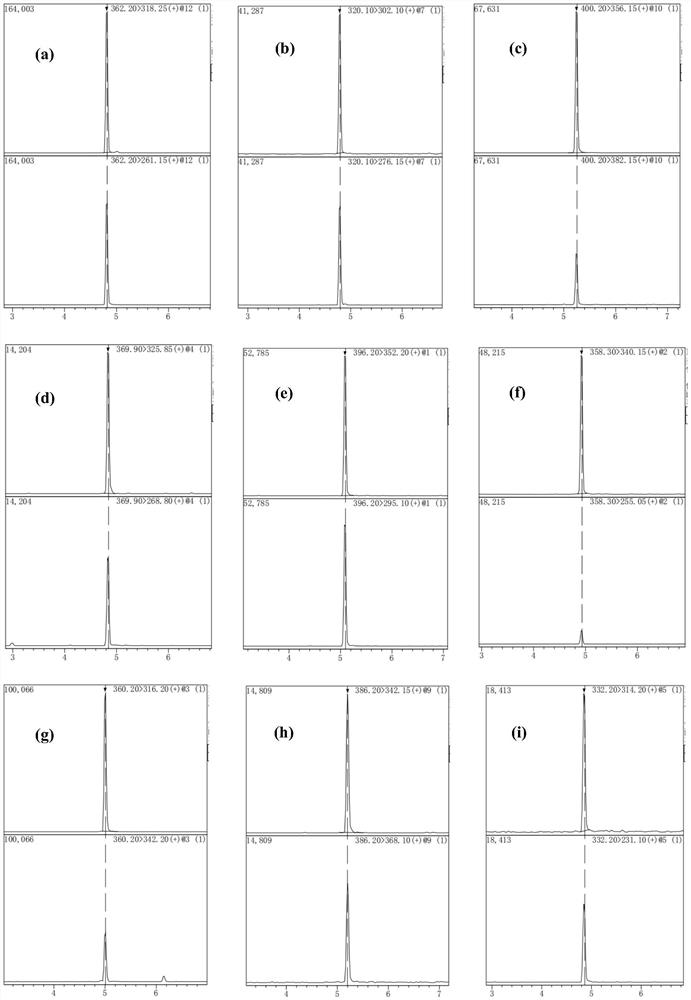
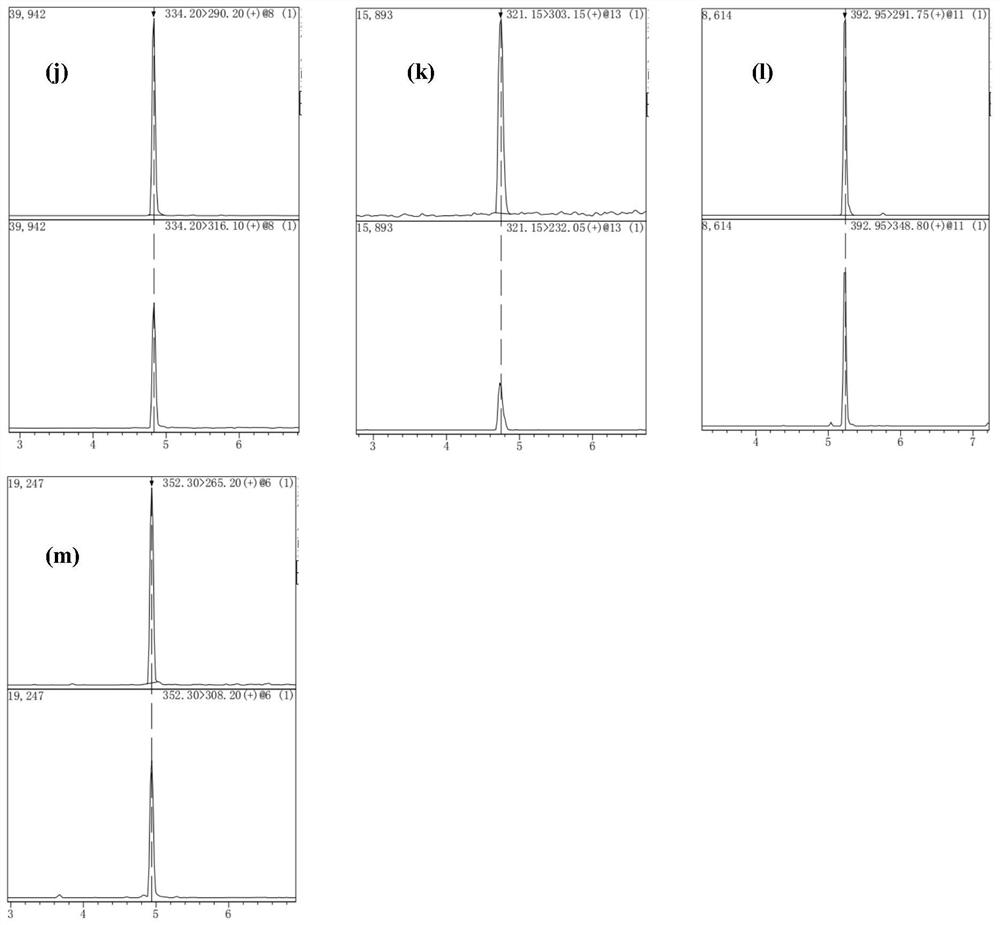
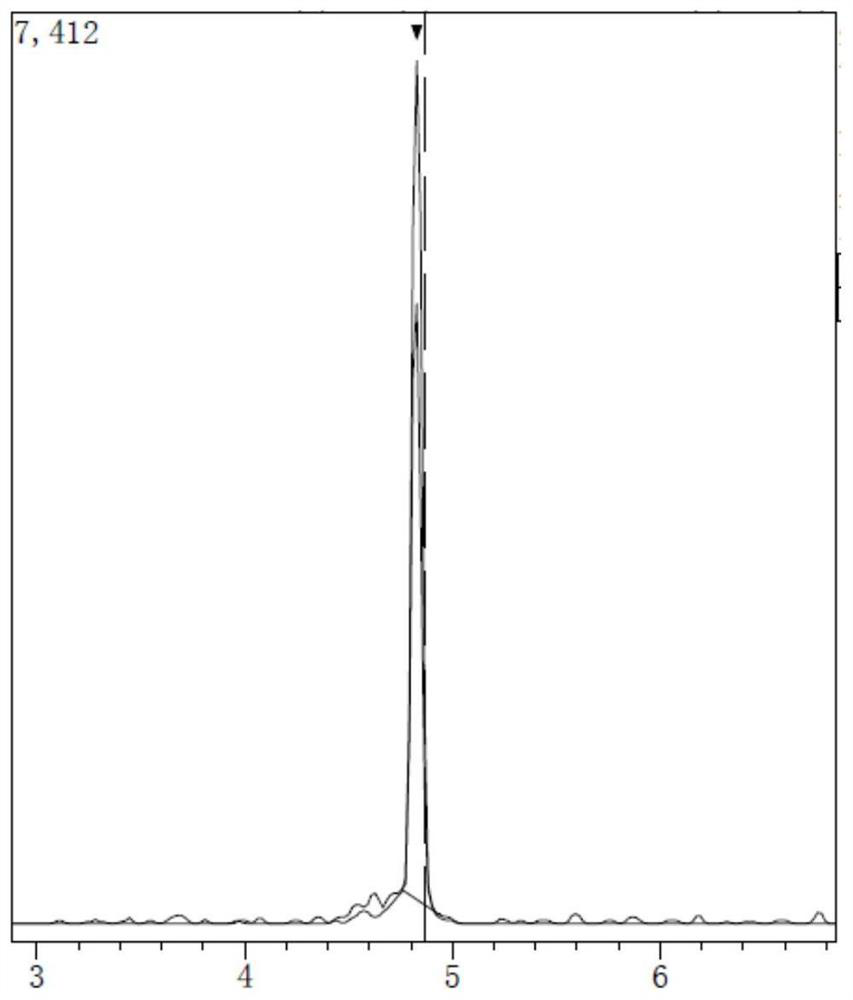
Method for detecting quinolones in feed
A technology of quinolones and detection methods, which is applied in the field of feed detection, can solve the problems of insufficient sensitivity and accuracy, and cannot meet the detection requirements of quinolones, and achieve improved sensitivity and accuracy, good selectivity and sensitivity, and accurate qualitative improvement effect of ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Sample pretreatment method: Weigh 2.00 g of the feed to be tested (crushed through a 100-mesh sieve) into a 10 mL centrifuge tube, add 20 mL of acetonitrile-water solution (4:1, V / V) containing 0.1 wt% formic acid, and vortex mix , shake and extract for 10 min, take 1 mL of the supernatant, dilute to 10 mL with 10 wt % acetonitrile aqueous solution, centrifuge for 15 min, the supernatant is the extract, and carry out solid-phase extraction on the molecularly imprinted monolithic column on the extract, the extraction method is as follows:
[0057] Activate the molecularly imprinted monolithic column with methanol and water at a flow rate of 0.75mL / min for 4min, take 1mL of the extract and load the sample, rinse the column with water at a flow rate of 0.75mL / min for 4min, and then use a volume ratio of 1.5:1 The methanol / acetic acid eluent was eluted for 8 minutes, and the eluate was collected. After blowing dry with nitrogen at 45°C, the residue was dissolved in 1 mL of 0...
Embodiment 2
[0066] Sample pretreatment method: Weigh 1.00 g of the feed to be tested (crushed through a 100-mesh sieve) into a 10 mL centrifuge tube, add 15 mL of acetonitrile-water solution (5:1, V / V) containing 0.15 wt% formic acid, and vortex mix , shake and extract for 12 minutes, take 1 mL of the supernatant, dilute to 10 mL with 10wt% acetonitrile aqueous solution, and centrifuge for 12 minutes.
[0067] Activate the molecularly imprinted monolithic column with methanol and water at a flow rate of 0.5mL / min in sequence for 6min, take 1mL of the extract and load the sample, rinse the column with water at a flow rate of 0.5mL / min for 6min, and then use a volume ratio of 1:1 The methanol / acetic acid eluent was eluted for 10 minutes, and the eluate was collected. After drying with nitrogen at 40°C, the residue was dissolved in 0.5 mL of 0.15 wt% formic acid acetonitrile solution, and passed through a 0.22 μm filter membrane to obtain the pretreated sample;
[0068] The preparation meth...
Embodiment 3
[0076] Sample pretreatment method: Weigh 2.00 g of the feed to be tested (crushed through a 100-mesh sieve) into a 10 mL centrifuge tube, add 20 mL of acetonitrile-water solution (4:1, V / V) containing 0.15 wt% formic acid, and vortex mix , shake and extract for 15 minutes, take 1 mL of the supernatant, dilute to 10 mL with 10 wt% acetonitrile aqueous solution, centrifuge for 15 minutes, the supernatant is the extract, and carry out solid-phase extraction on the molecularly imprinted monolithic column on the extract, the extraction method is as follows:
[0077] Activate the molecularly imprinted monolithic column with methanol and water respectively at a flow rate of 1.0mL / min for 3min, take 0.5mL of the extract and load the sample, rinse the column with water at a flow rate of 1.0mL / min for 3min, and then use a volume ratio of 1: 1 methanol / acetic acid eluent was eluted for 5 minutes, the eluate was collected, and after being blown dry with nitrogen at 50°C, the residue was di...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap