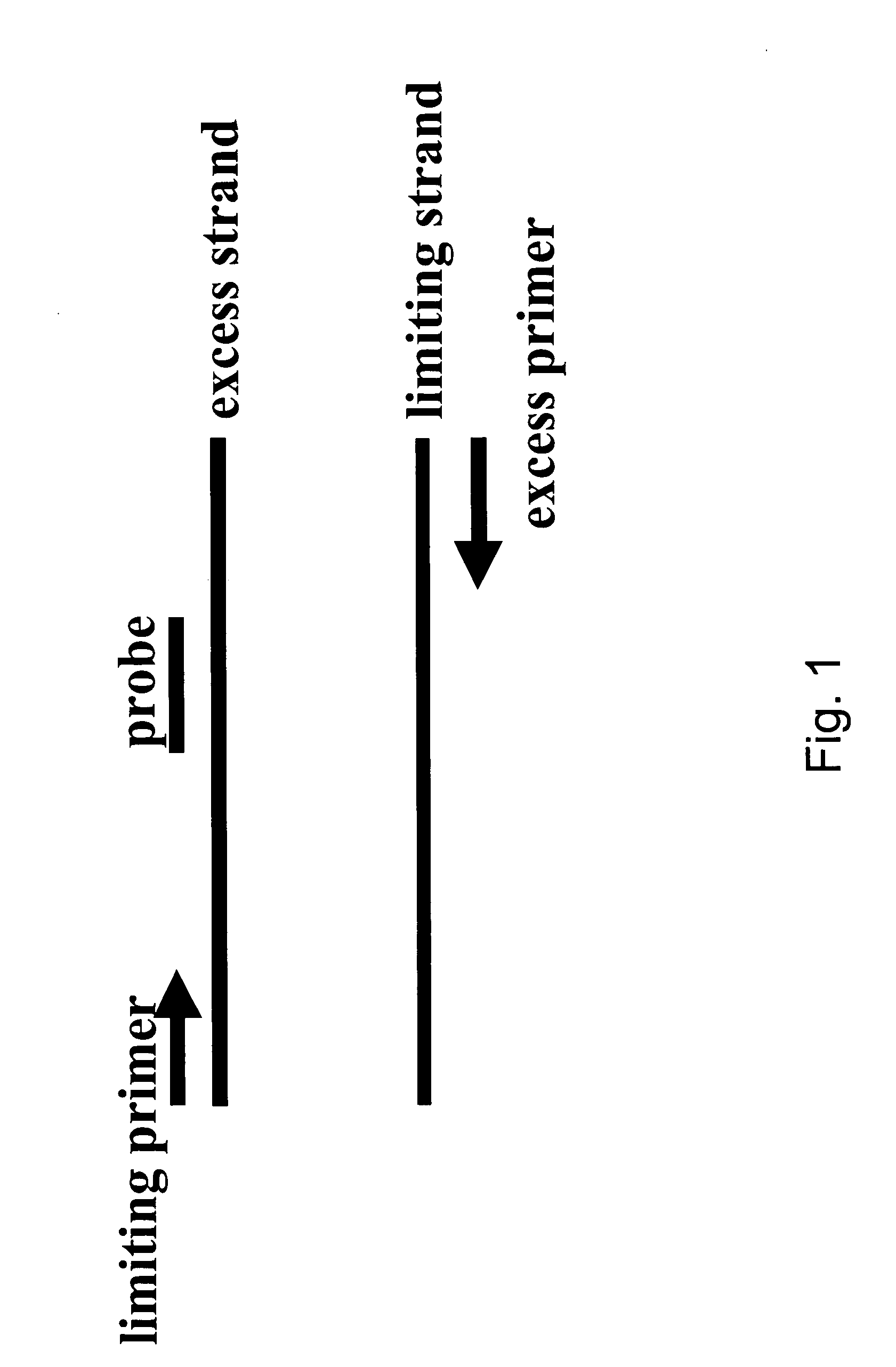
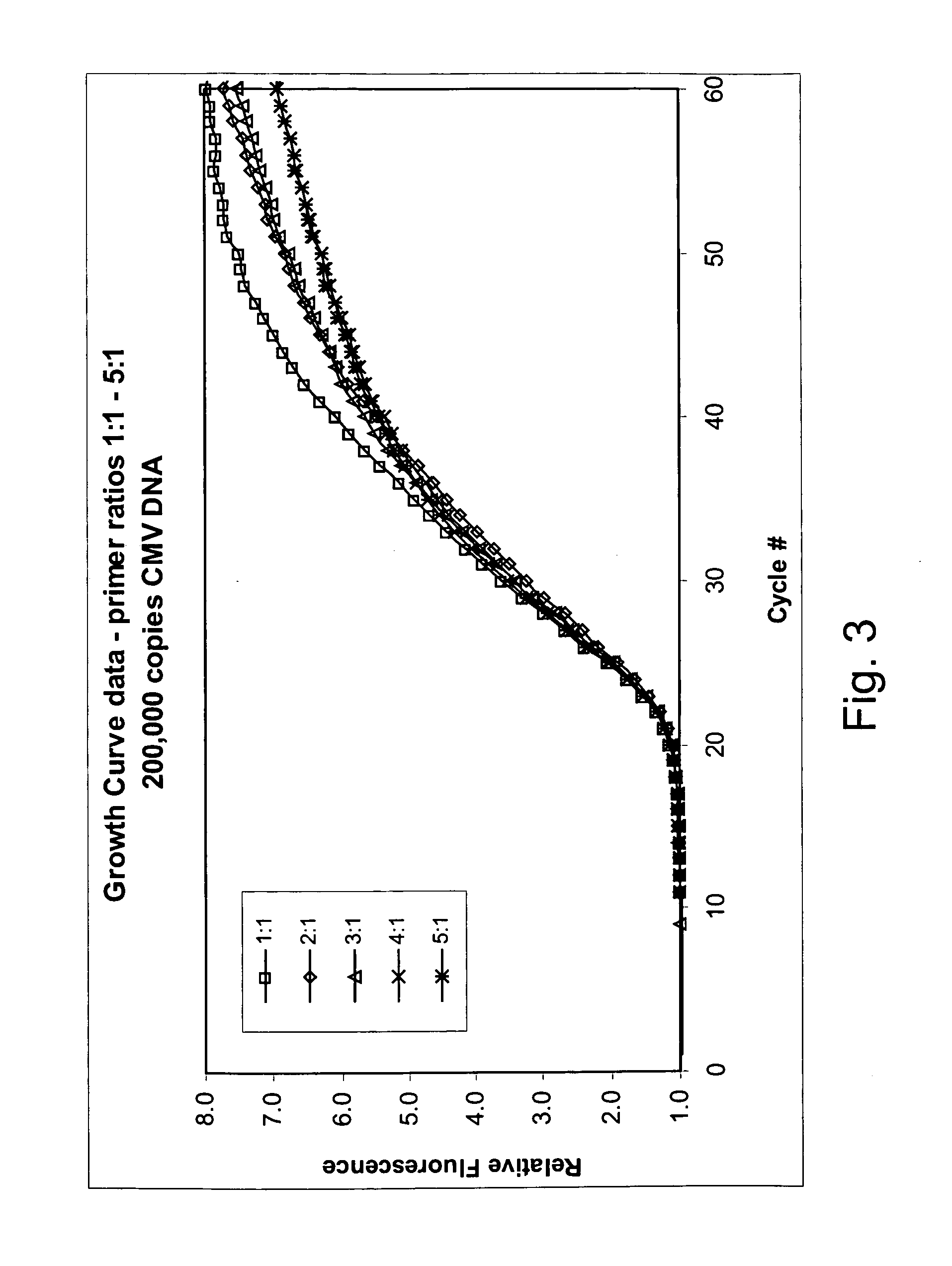
Asymmetric PCR coupled with post-PCR characterization for the identification of nucleic acids
a nucleic acid and characterization technology, applied in the field of nucleic acid chemistry and nucleic acid identification, can solve problems such as more difficult to solve, and achieve the effect of increasing intensity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
Use of the Invention to Determine HCV Genotype
[0119] Determining the genotype of an HCV (hepatitis C virus) positive sample has important clinical significance and utility. Knowing the genotype of the specific virus allows physicians to select the correct treatment regimen. As explained above, the current invention can be used to discriminate amongst a number of nucleic acid targets (e.g., even ones that are closely related in sequence) such as HCV genotypes. The methods of the invention use an oligonucleotide probe that is specifically designed to be at least partially complementary to a region of target sequence divergence (e.g., a region that shows divergence between different strains, alleles, species, etc.) in conjunction with using the different Tms obtained from post-PCR melting and / or annealing analysis to distinguish the nucleic acid targets.
[0120]FIG. 5 shows aligned sequences of 6 different HCV genotypes (namely types 1a, 2a, 3a, 4, 5, and 6) along with an exemplary hyb...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com