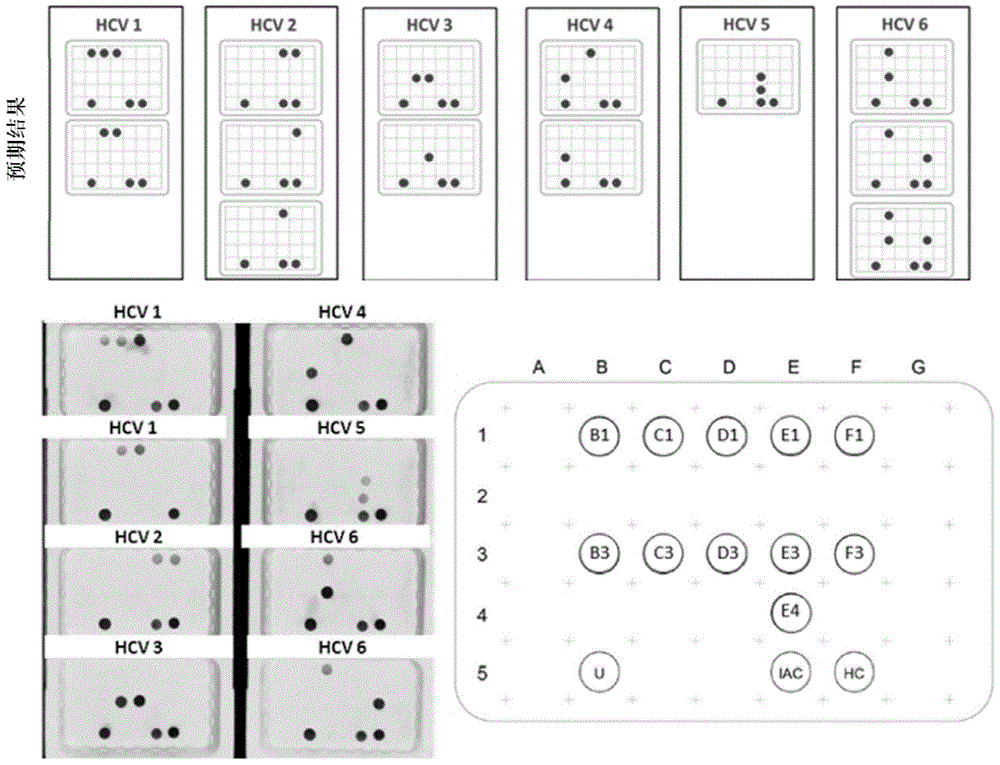
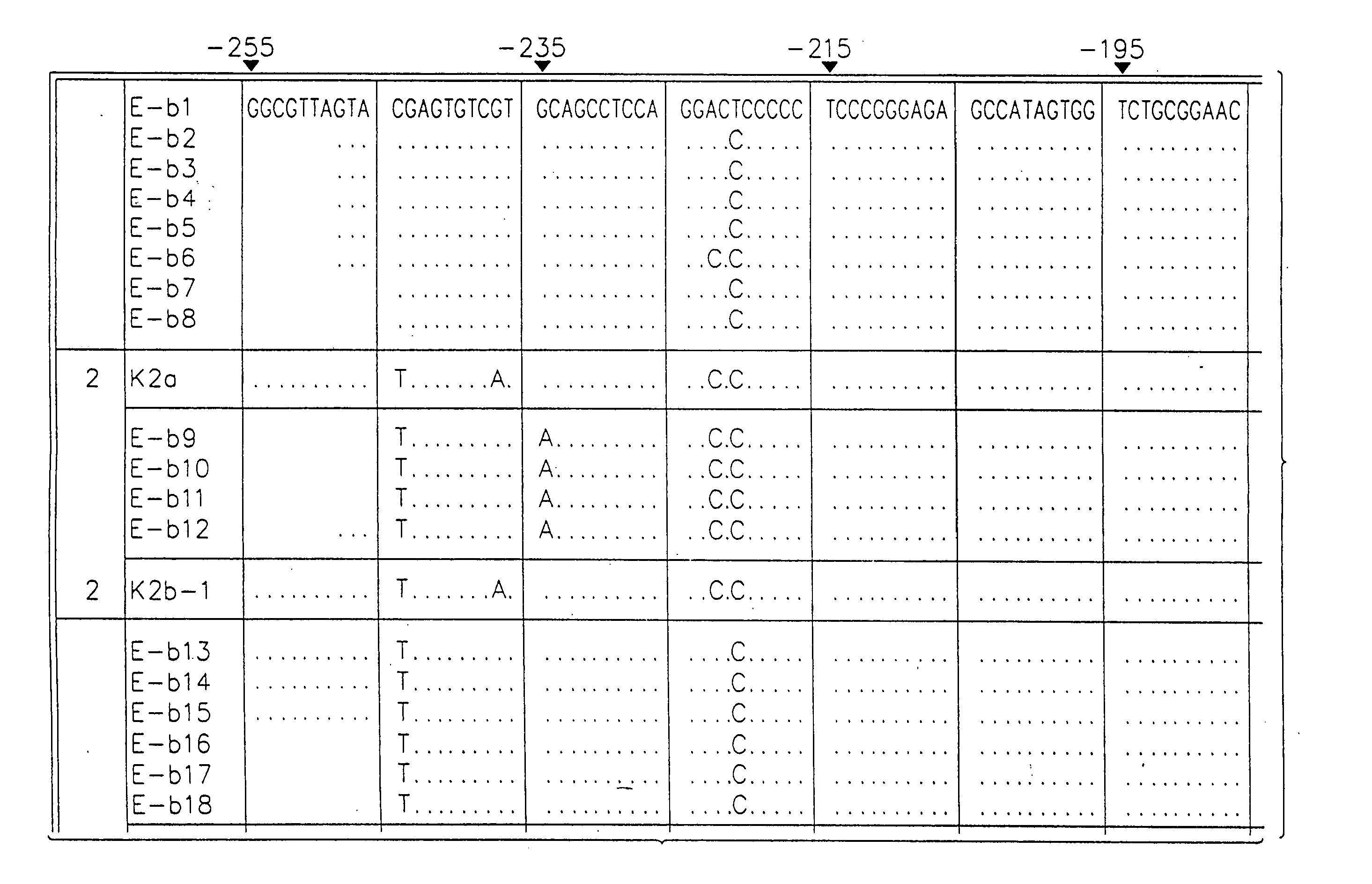
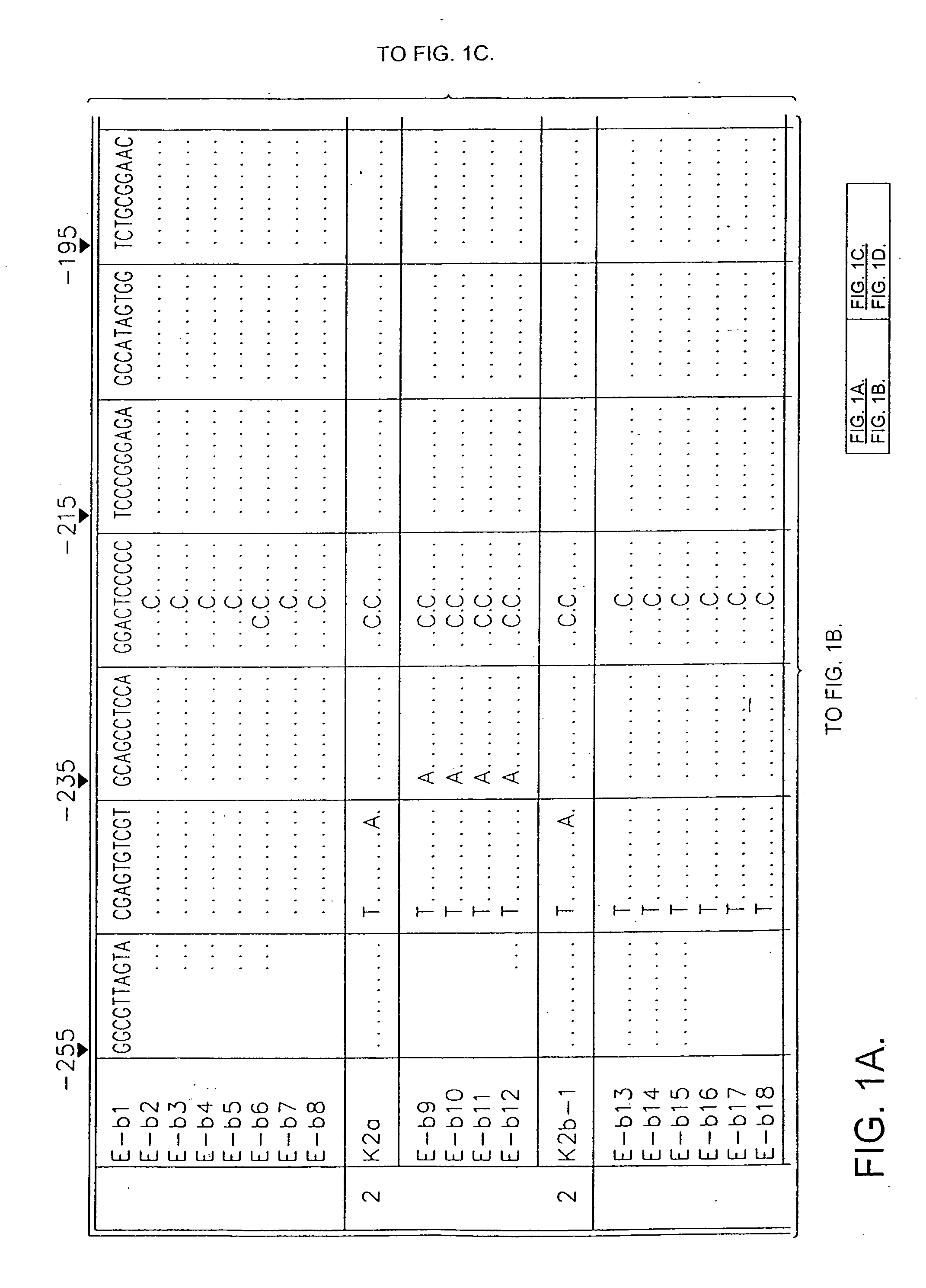
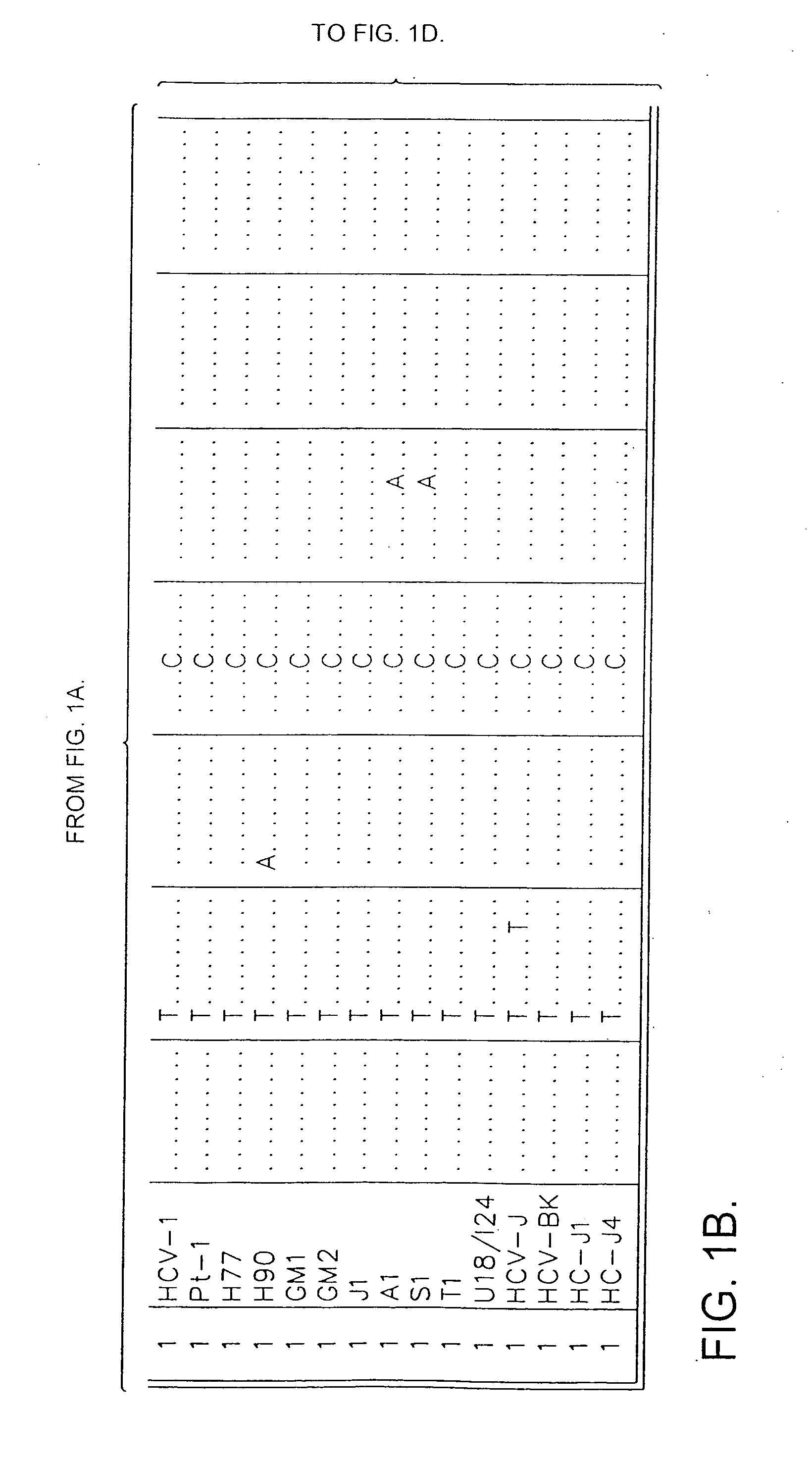
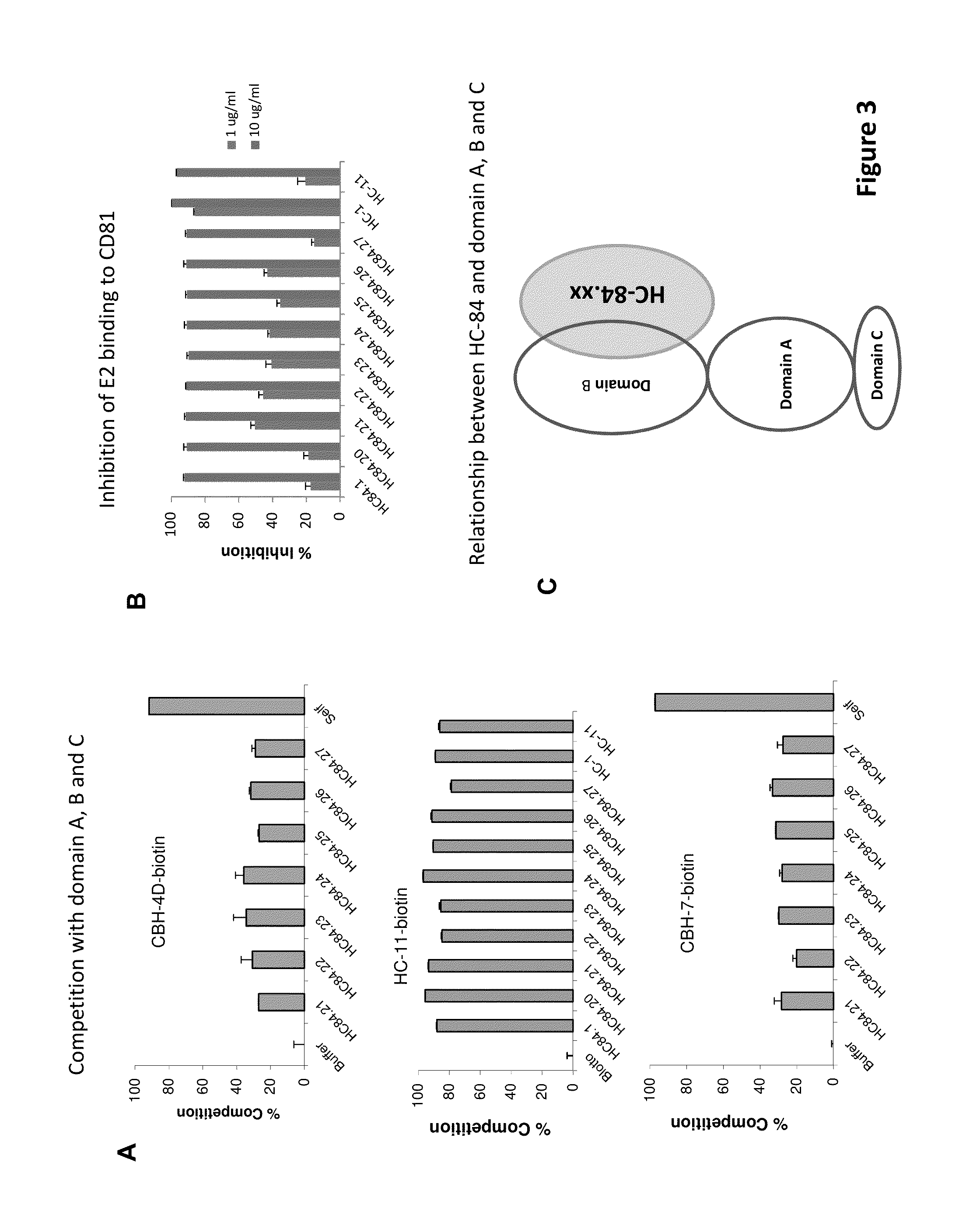
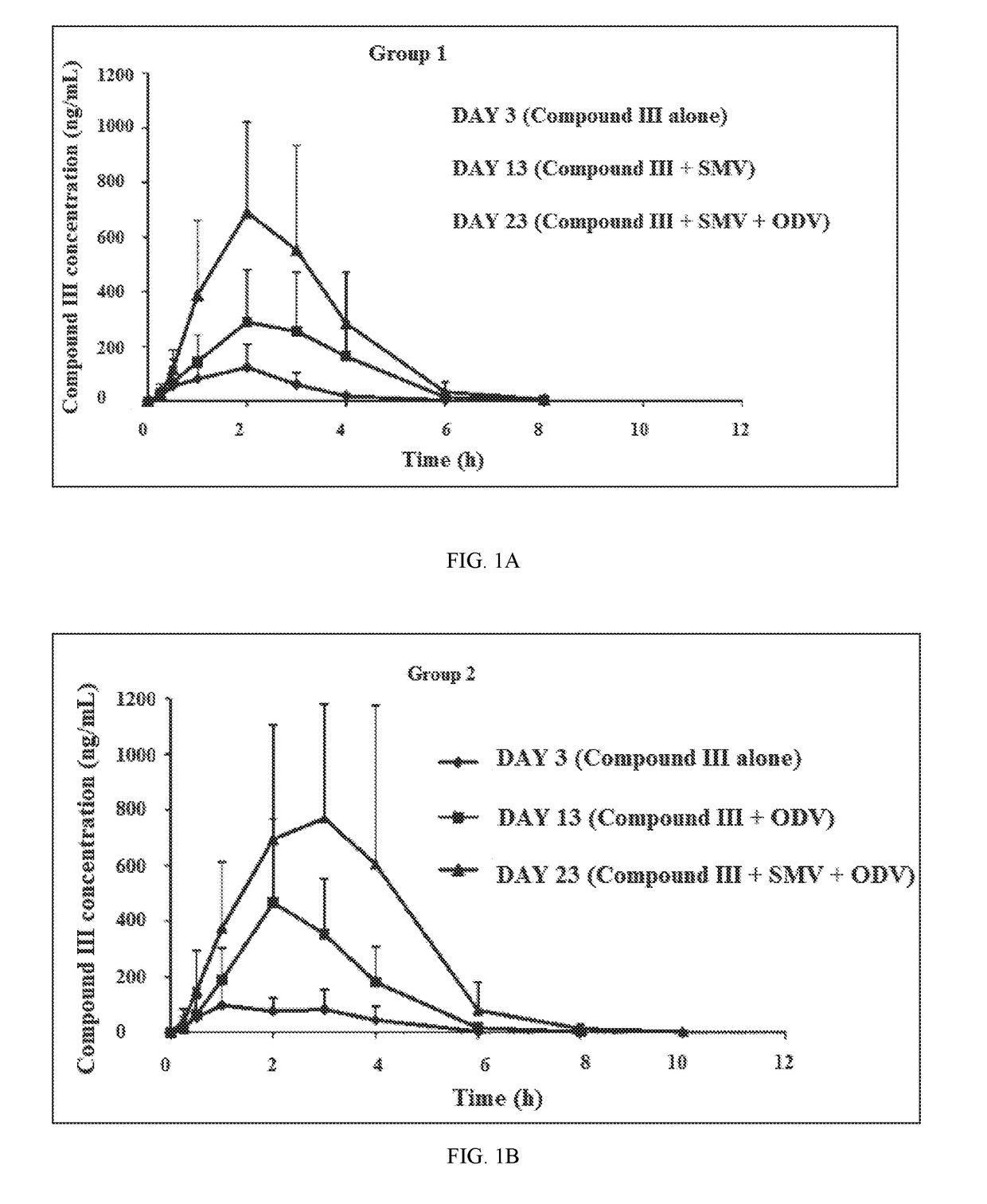
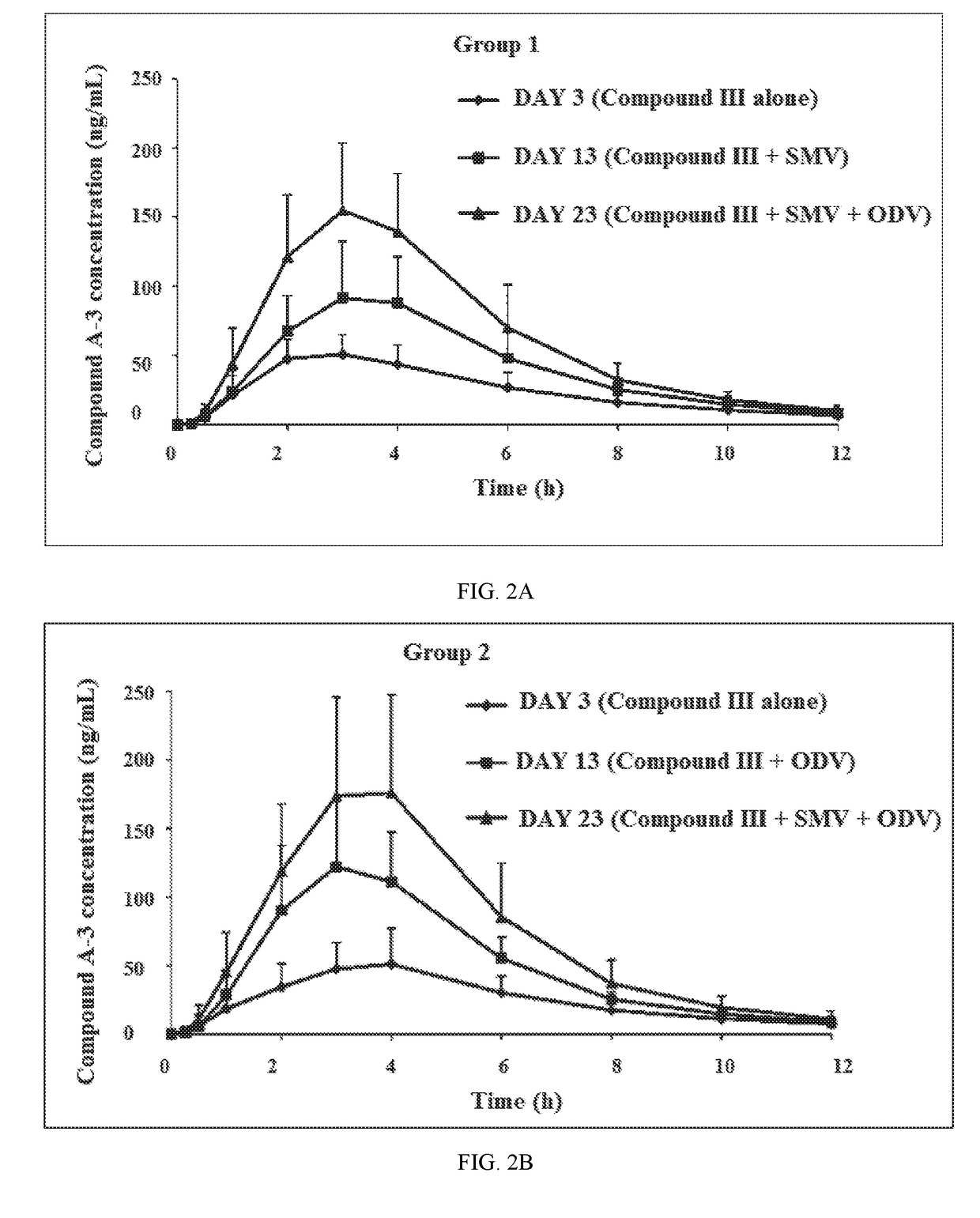
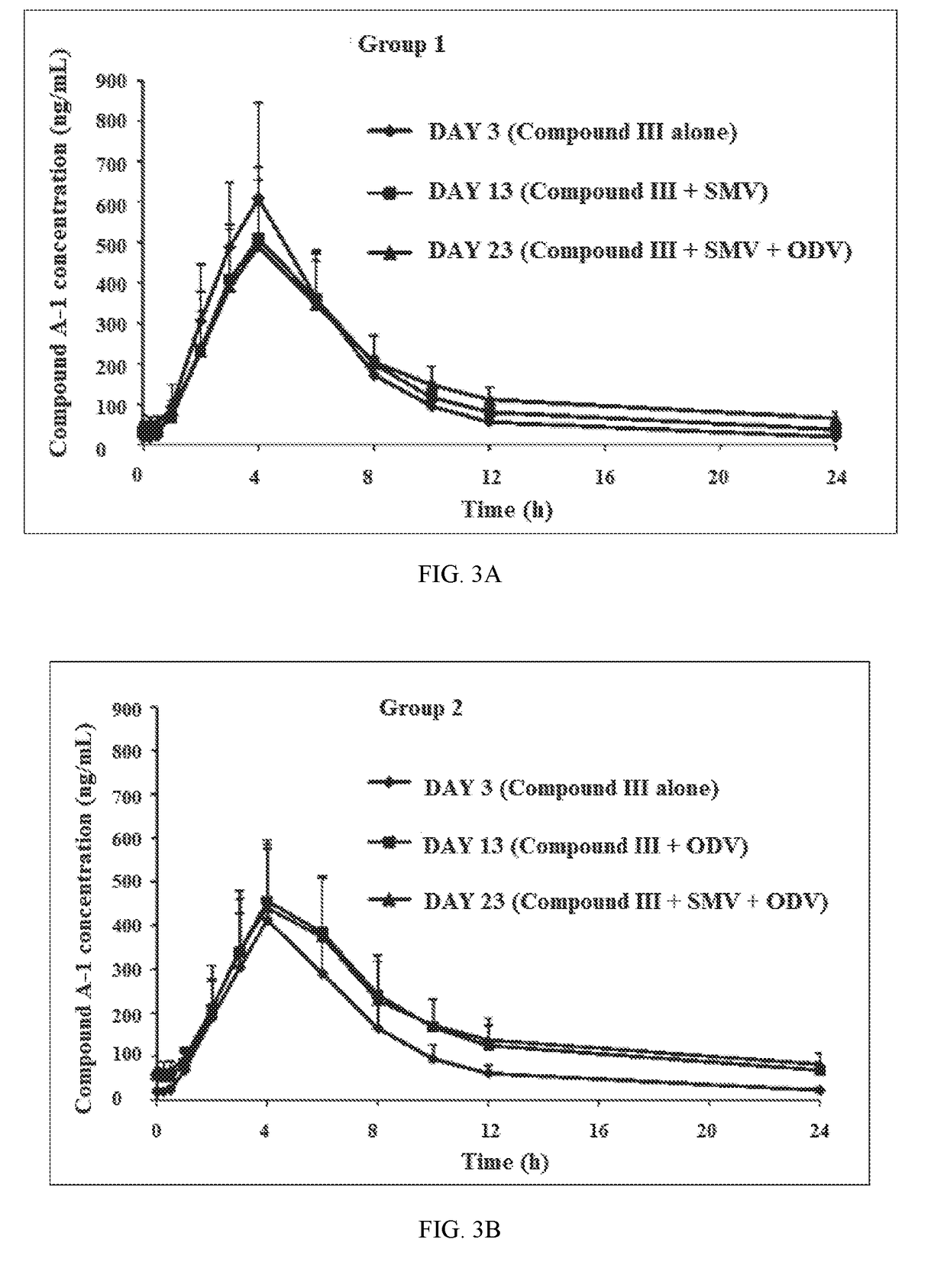
Patents
Literature
61 results about "HCV Genotyping" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Asymmetric PCR coupled with post-PCR characterization for the identification of nucleic acids
InactiveUS20070072211A1Microbiological testing/measurementEnzymologyHCV GenotypingNucleic acid sequencing
Owner:ROCHE MOLECULAR SYST INC
Hcv Ns3-Ns4a Protease Inhibition
InactiveUS20080267915A1Inhibit HCV replicationReduce riskDipeptide ingredientsTetrapeptide ingredientsHCV GenotypingProtease
The present invention relates to inhibiting the activity of non-genotype 1 hepatitis C virus (HCV) NS3-NS4A protease activity. More particularly, the invention relates to inhibiting the activity of the protease from HCV genotype-2 or HCV genotype-3. The methods of the invention emply inhibitors that act by interfering with the life cycle of the HCV and are also useful as antiviral agents. The invention further relates to compositions comprising such compounds either for ex vivo use or for administration to a patient suffering from genotype-2 or genotype-3 HCV infection. The invention also relates to methods of treating an HCV infection in a patient by administering a composition comprising a compound of this invention.
Owner:VERTEX PHARMA INC
Probes and methods for hepatitis C virus typing using single probe analysis
InactiveUS20070020665A1Less stringentIncrease ionic strengthMicrobiological testing/measurementEnzymologyHCV GenotypingHcv hepatitis c virus
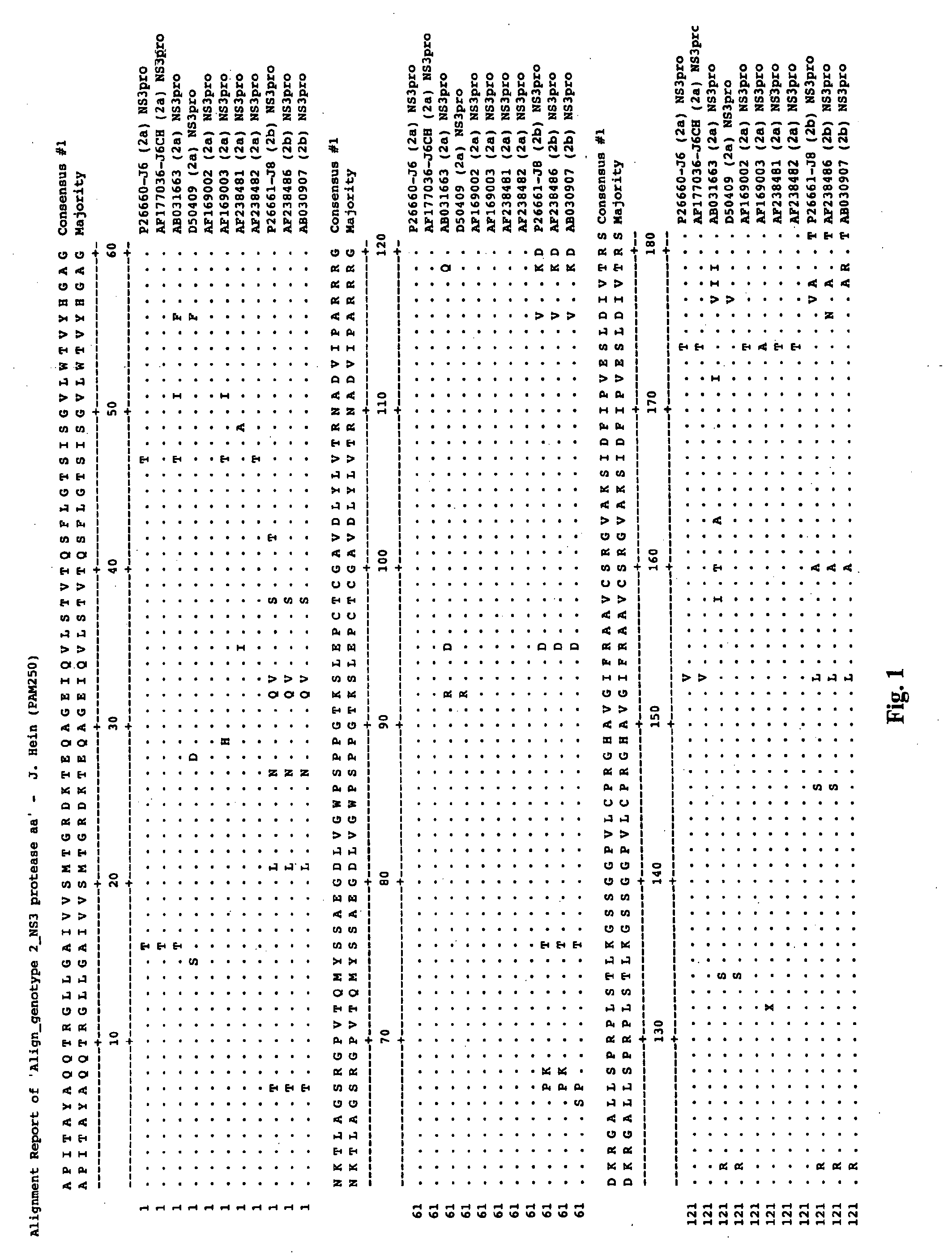
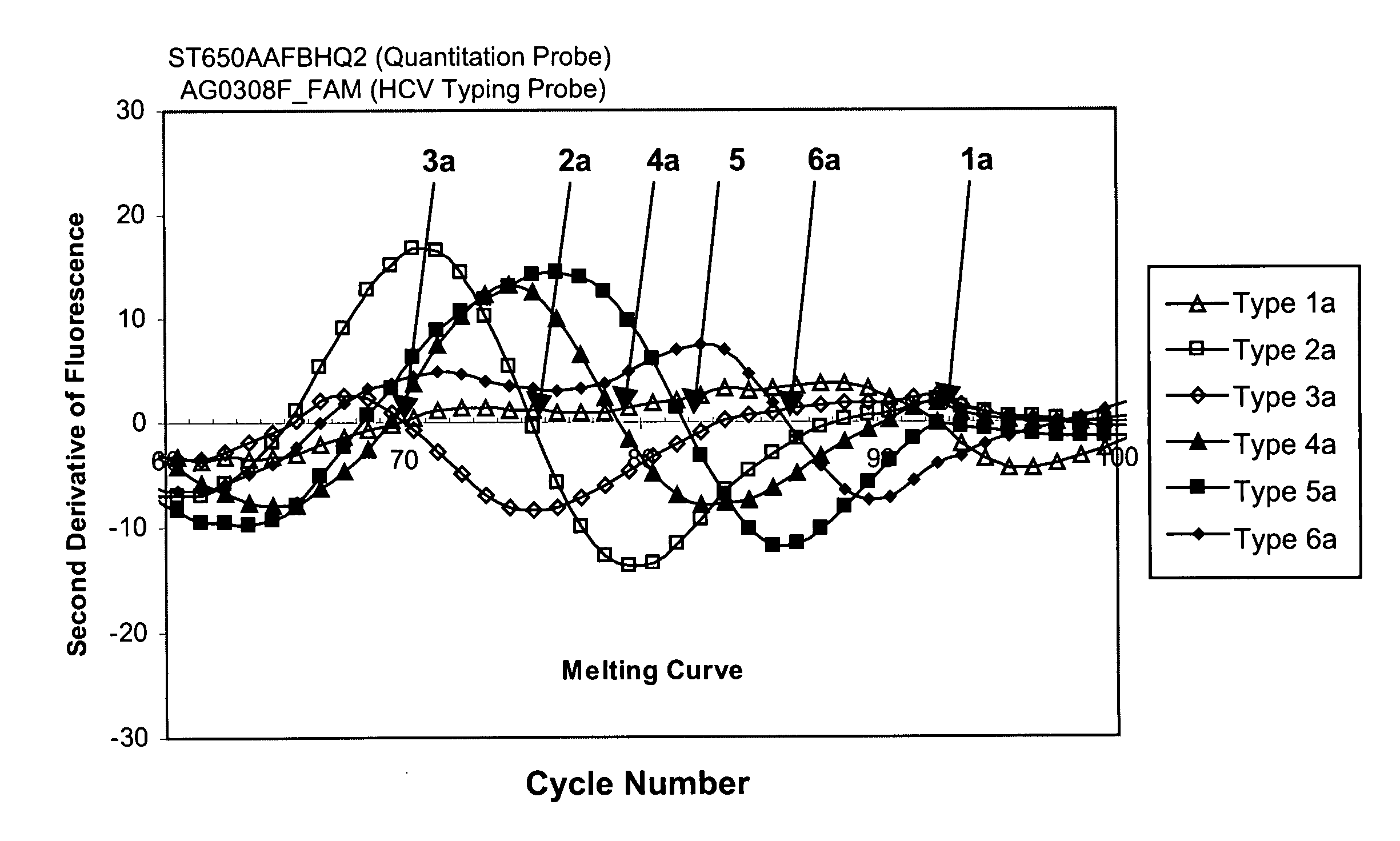
This invention provides compositions and methods for HCV typing, e.g., genotyping and / or subtyping. The compositions and methods of the invention can be used to assign an HCV isolate to one of at least five HCV genotypes (for example, selected from genotypes 1, 2, 3, 4, 5 or 6), or assign an HCV isolate to one of at least six subtypes (for example, selected from subtypes 1a / b / c, 2a / c, 2b, 3a, 4a, 5a or 6a), where the methods of the invention use only a single typing probe to make the HCV type assignment.
Owner:ROCHE MOLECULAR SYST INC
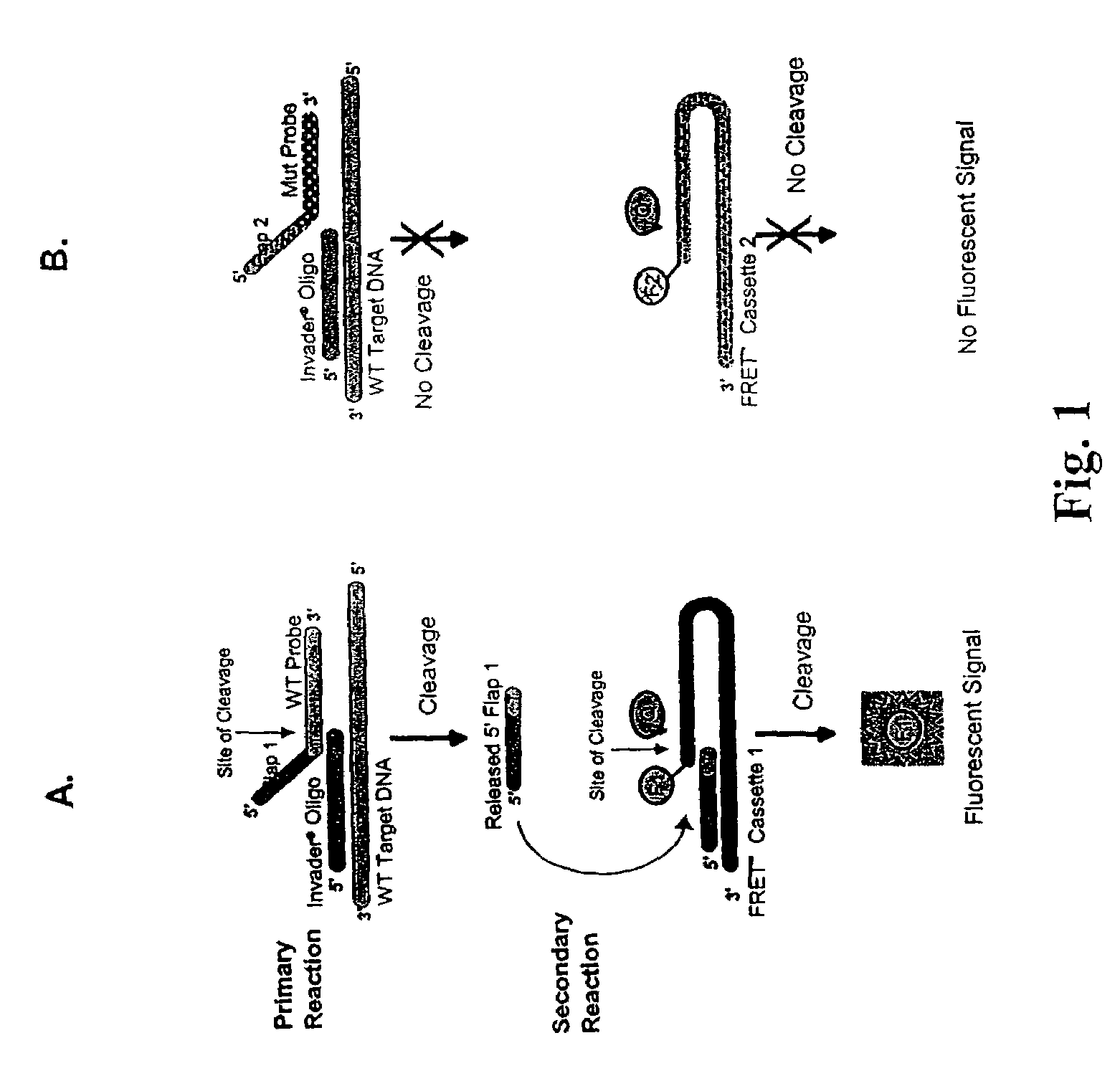
Determination of hepatitis C virus genotype
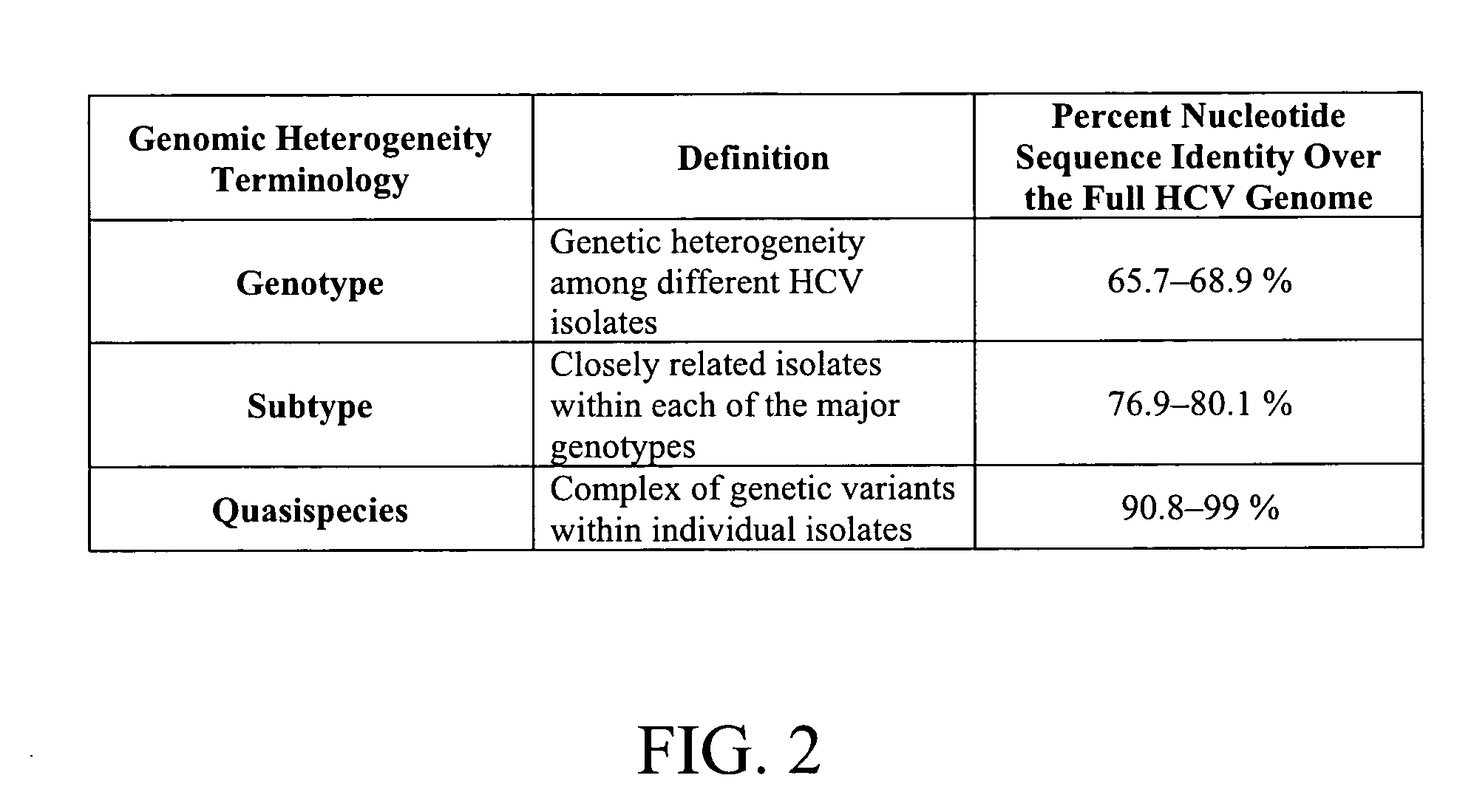
The present invention provides compositions and methods for the detection and characterization of HCV sequences. More particularly, the present invention provides compositions, methods and kits for using invasive cleavage structure assays (e.g. the INVADER assay) to screen nucleic acid samples, e.g., from patients, to determine HCV genotype.
Owner:GEN PROBE INC
Efficient cell culture system for hepatitis C virus genotype 5A
Owner:HVIDOVRE HOSPITAL
Cell culture system of a hepatitis c genotype 3a and 2a chimera
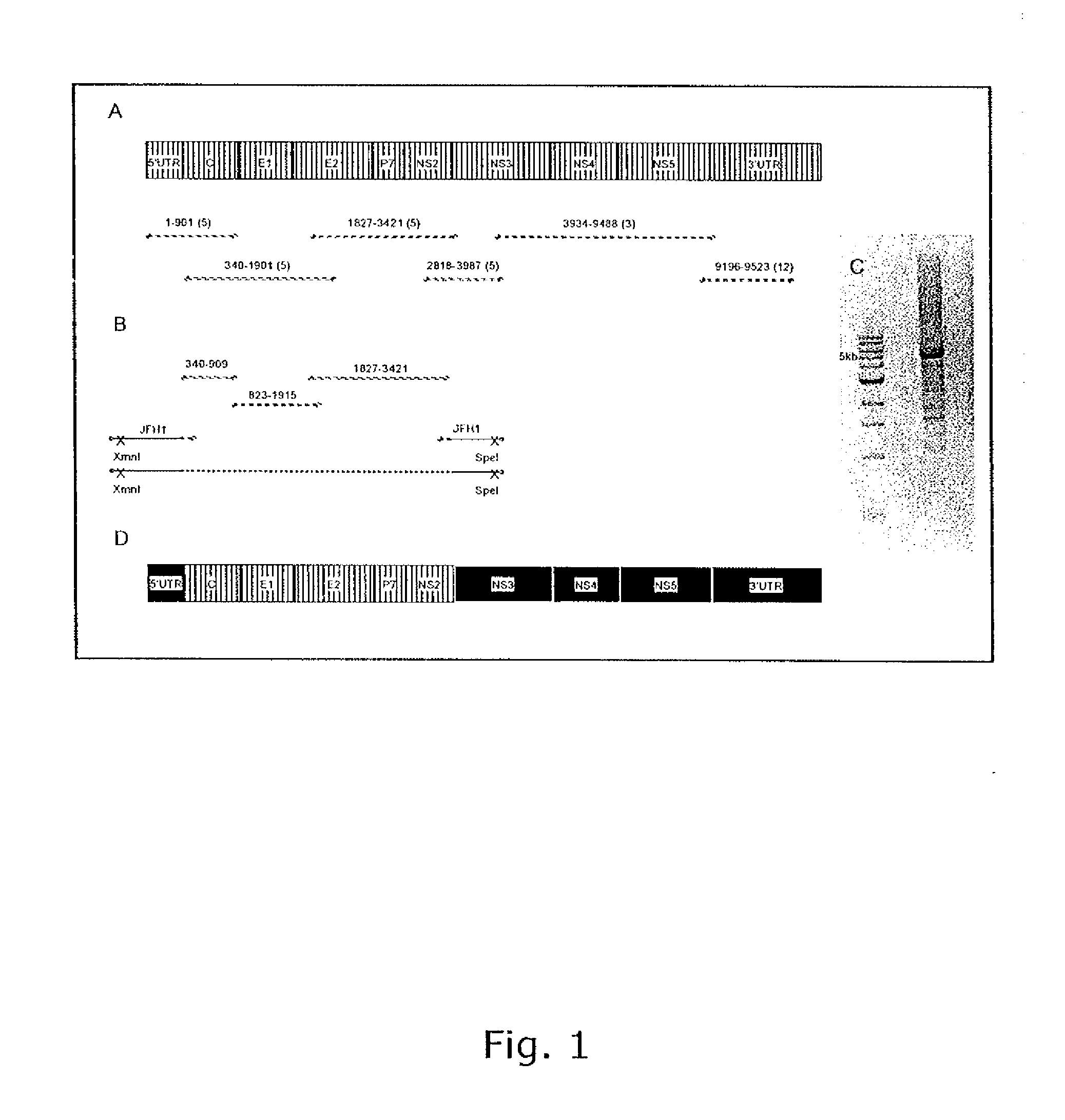
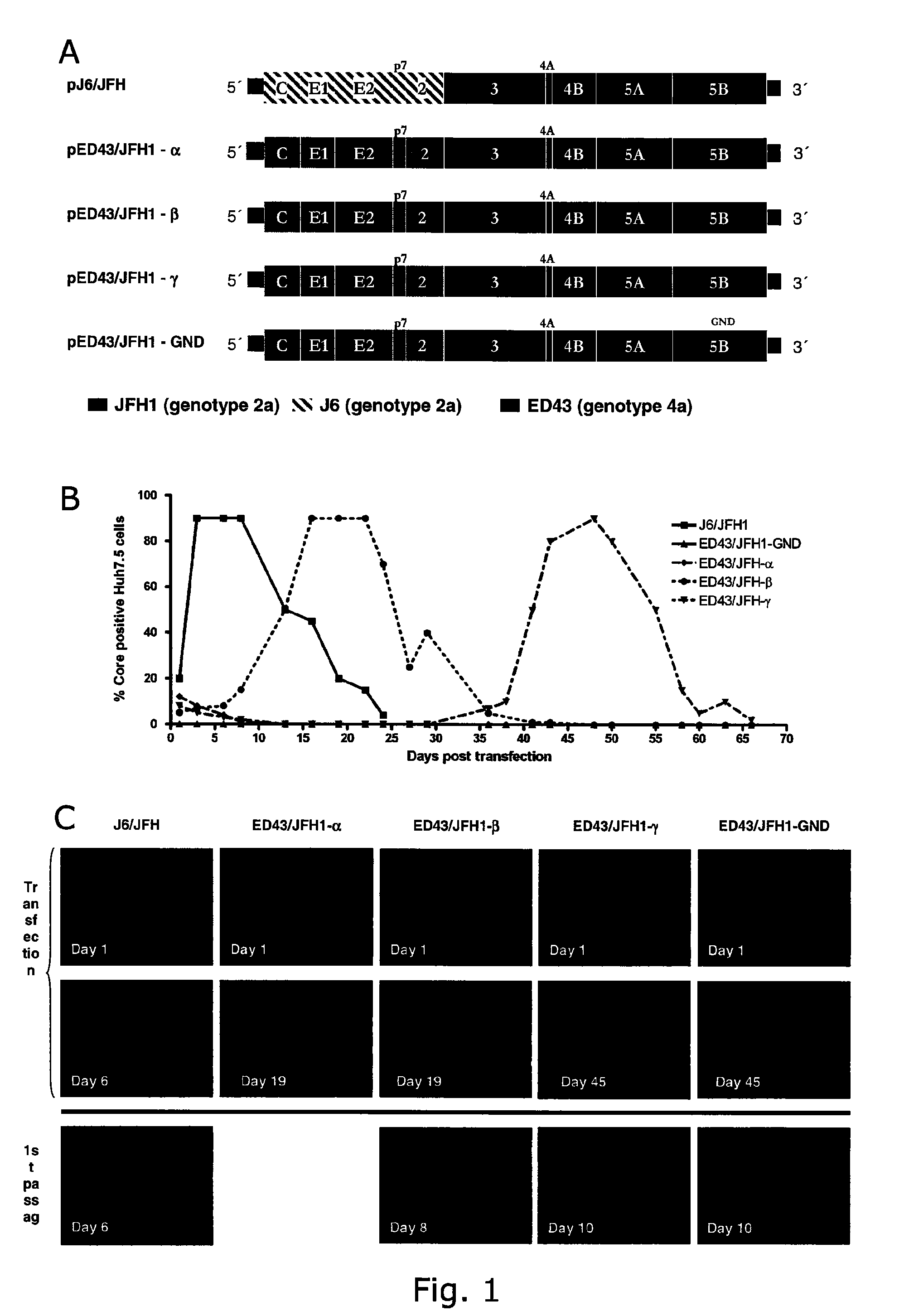
InactiveUS20100093841A1Efficient and sustainable growthDifferential efficiencyOrganic active ingredientsSsRNA viruses positive-senseGenomic sequencingNS5A
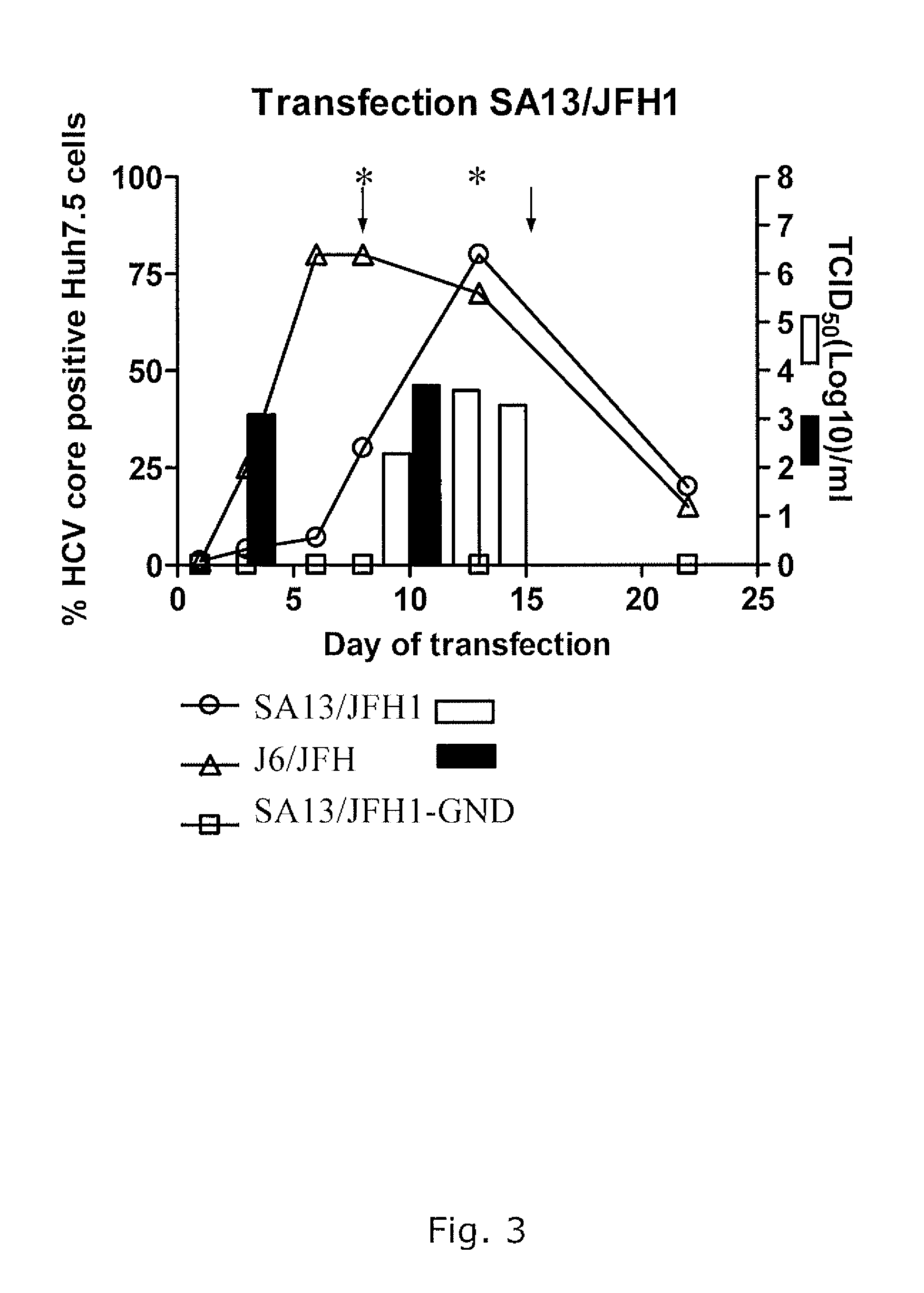
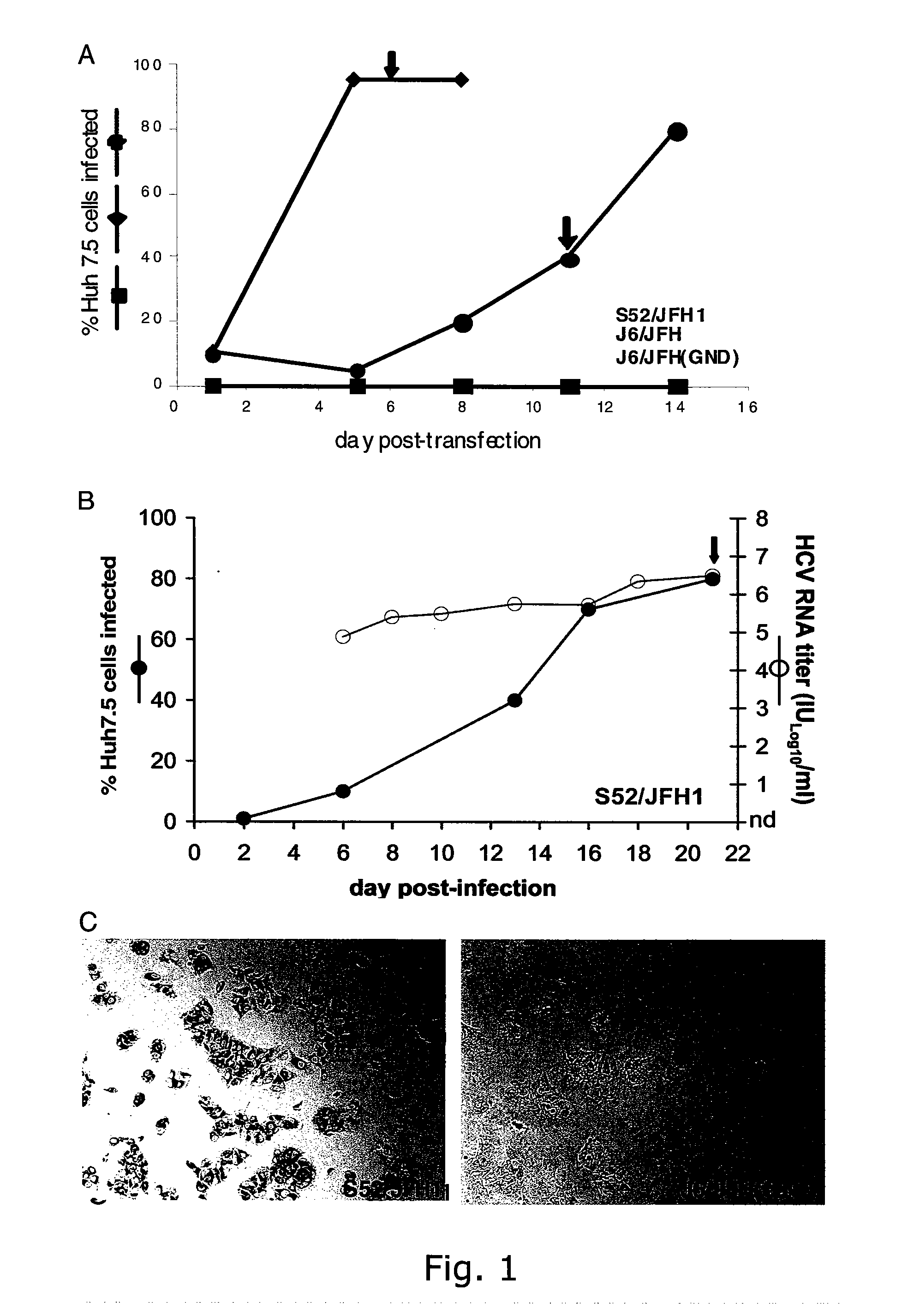
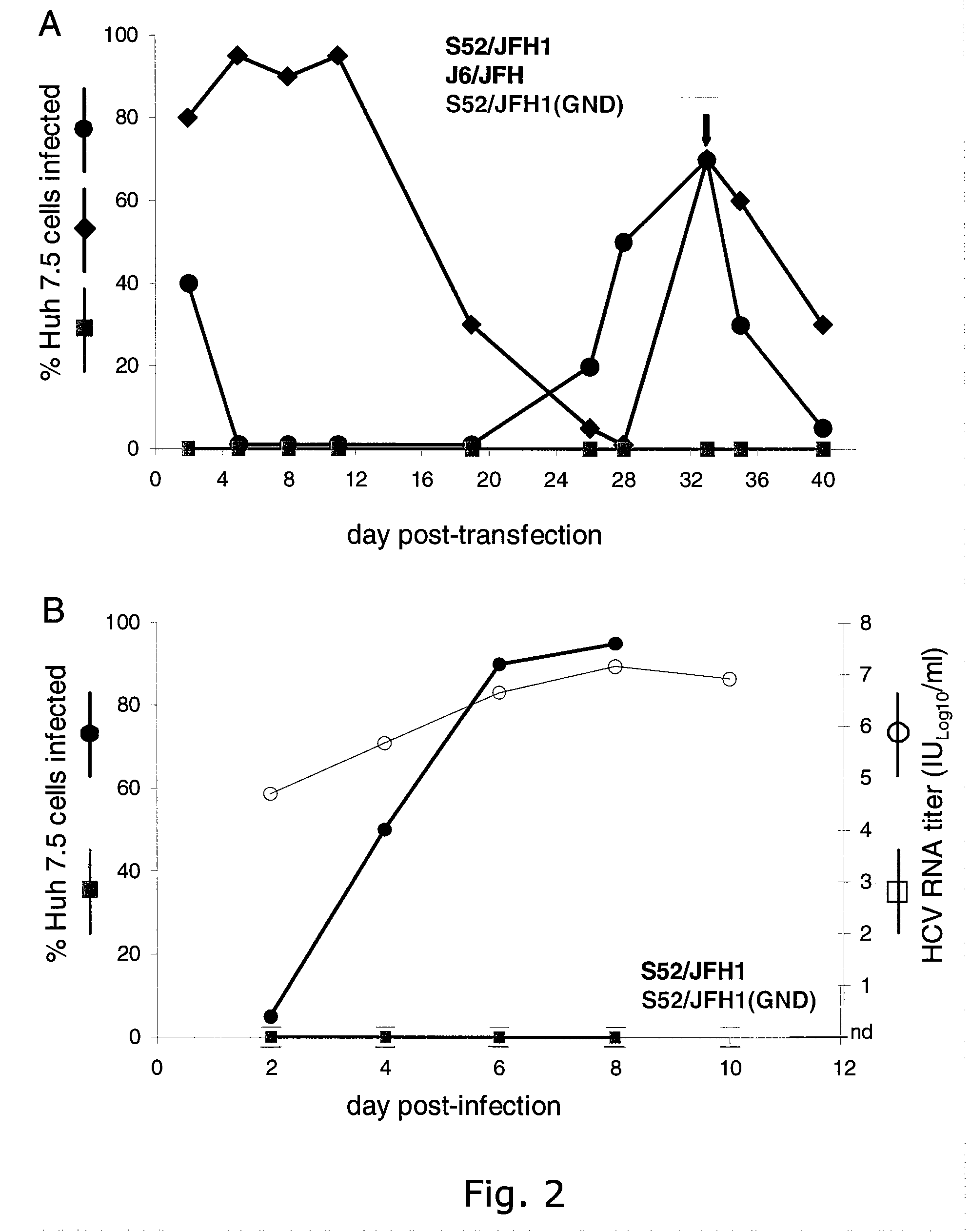
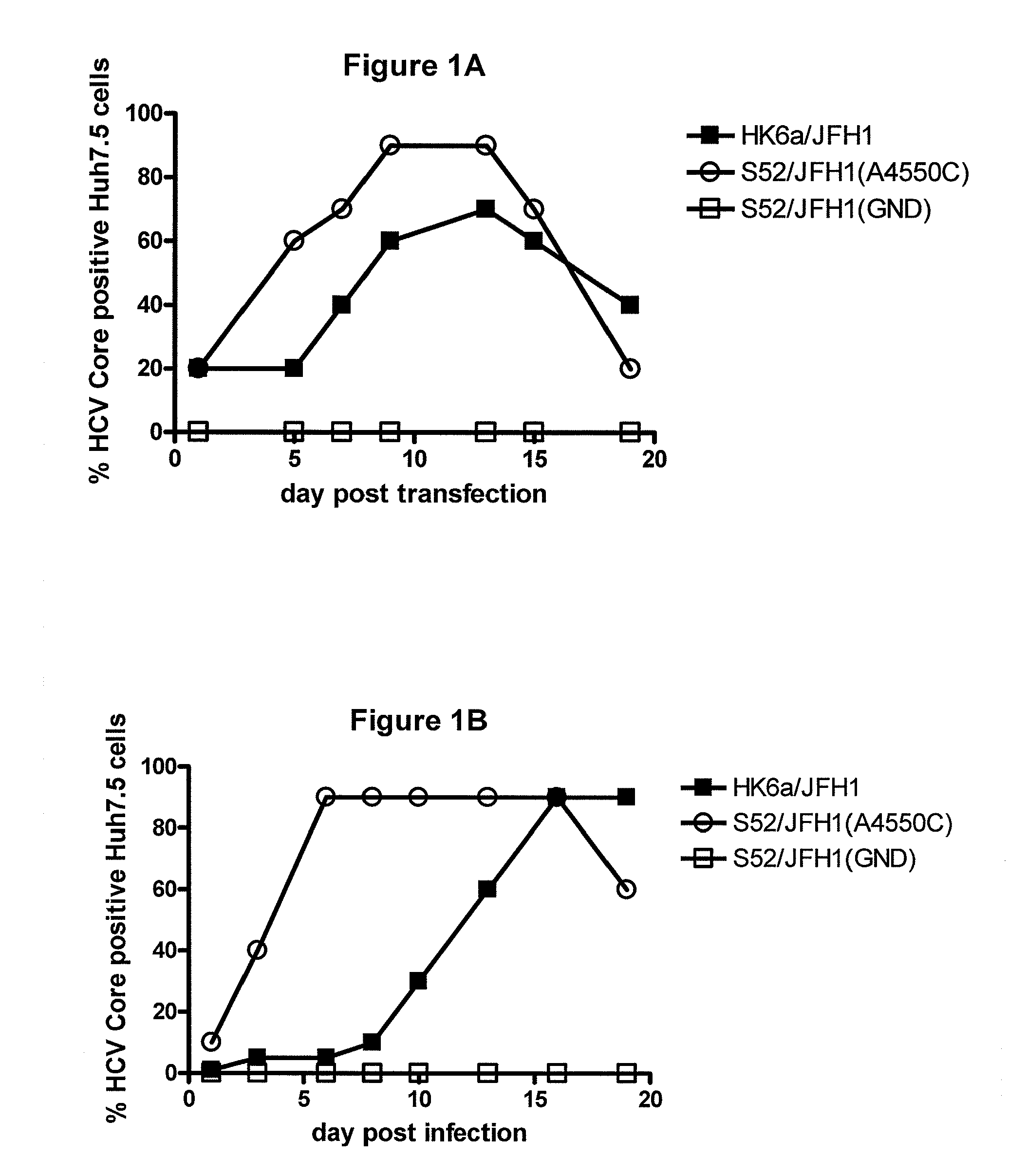
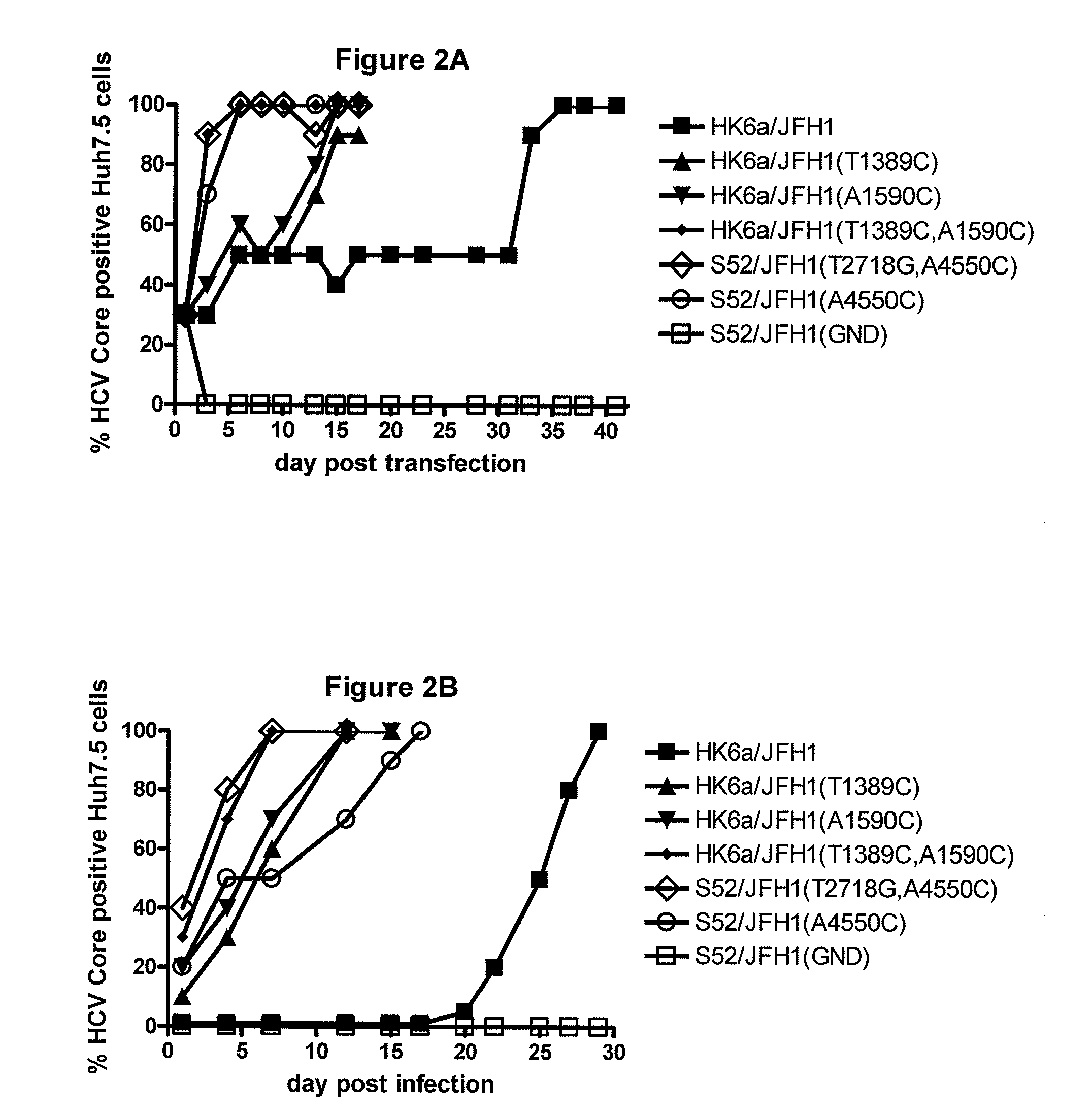
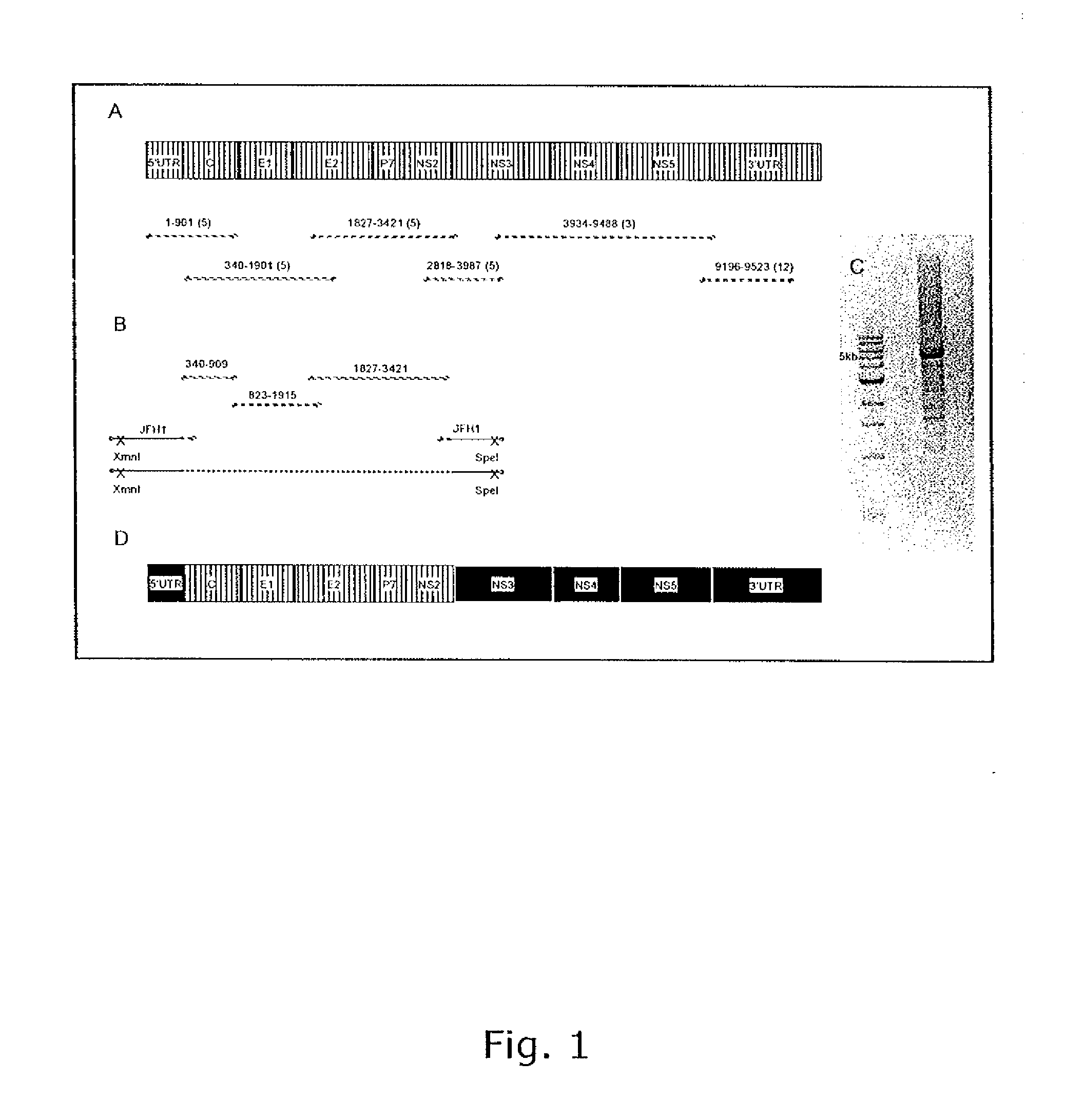
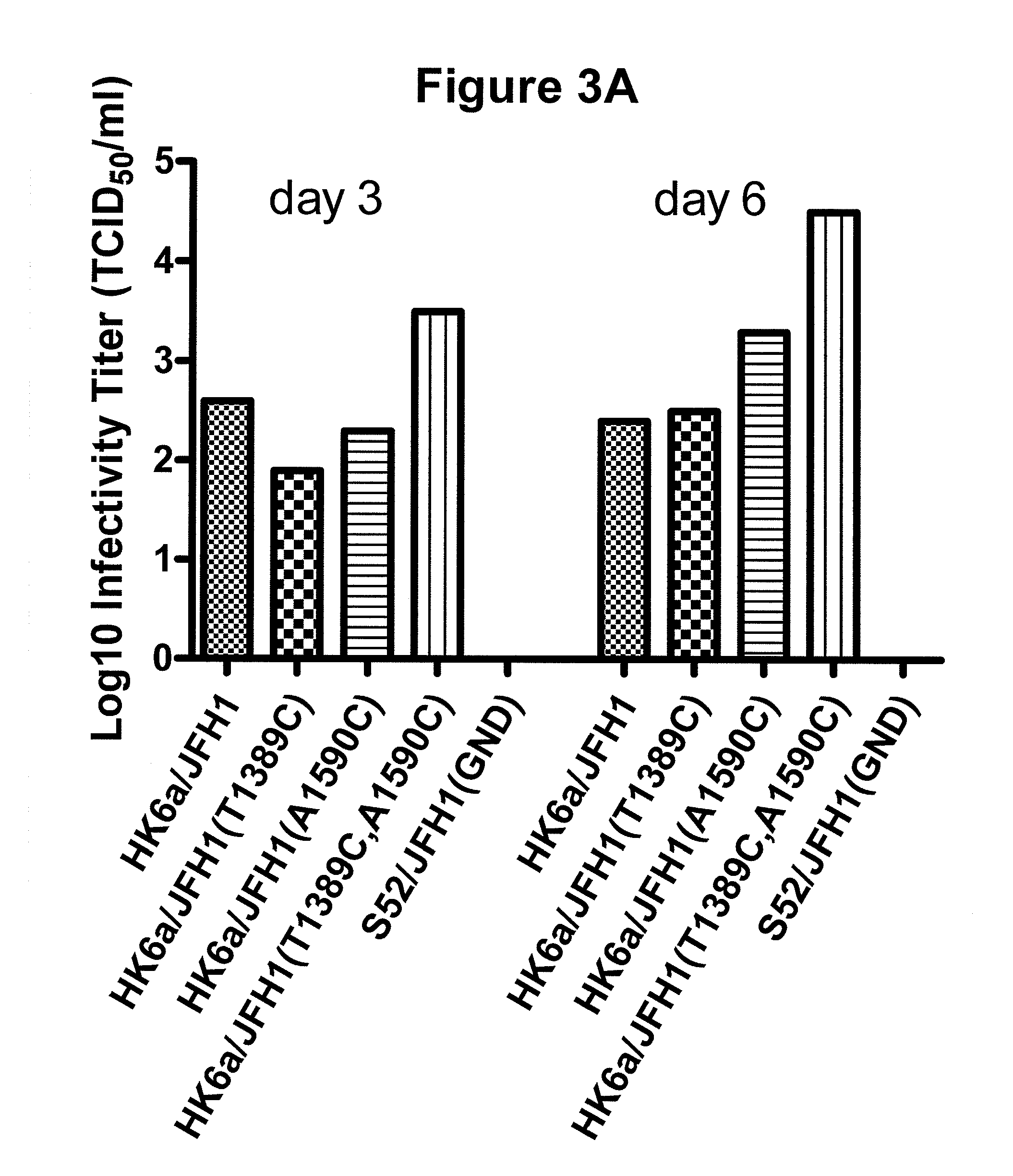
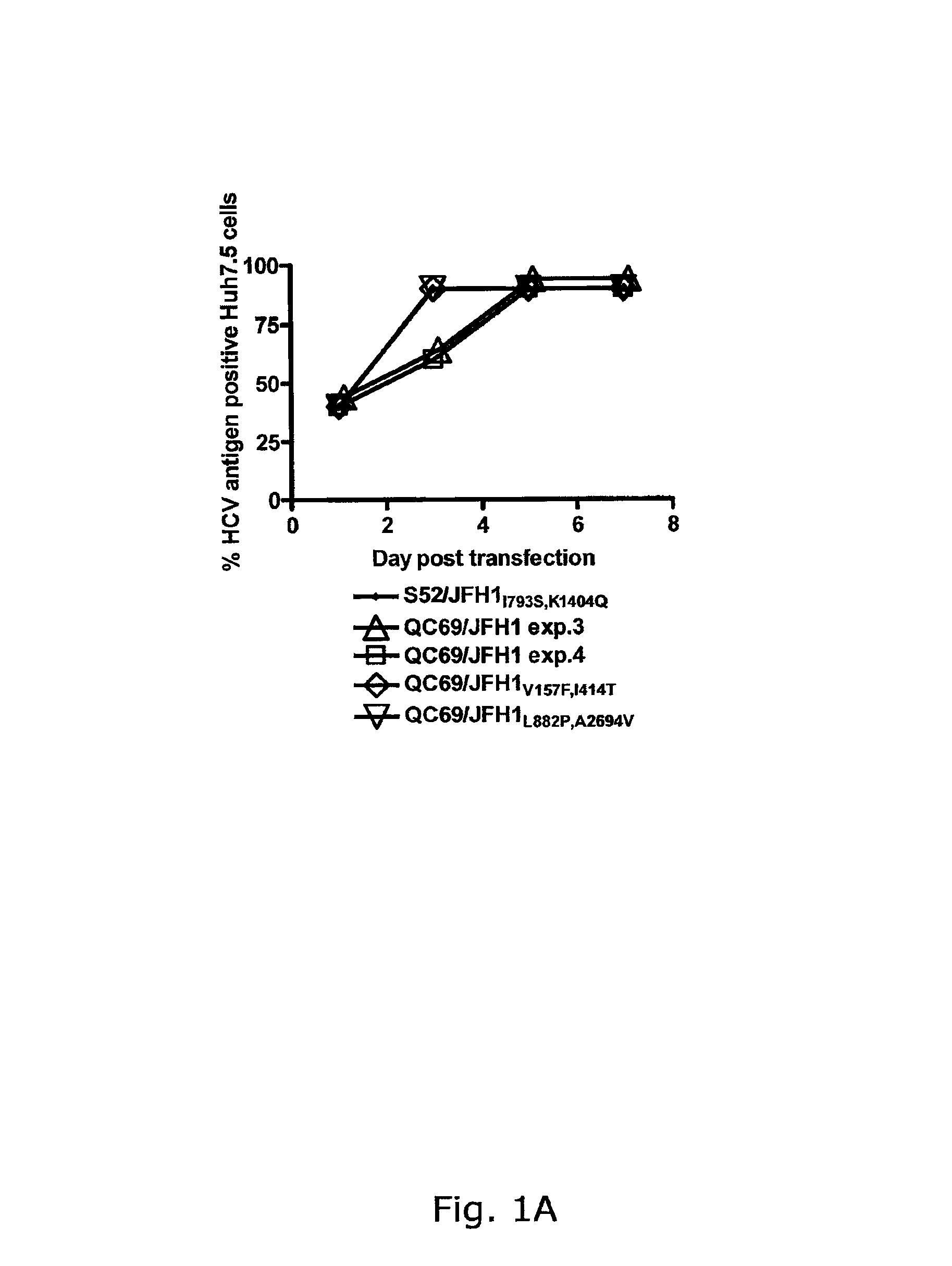
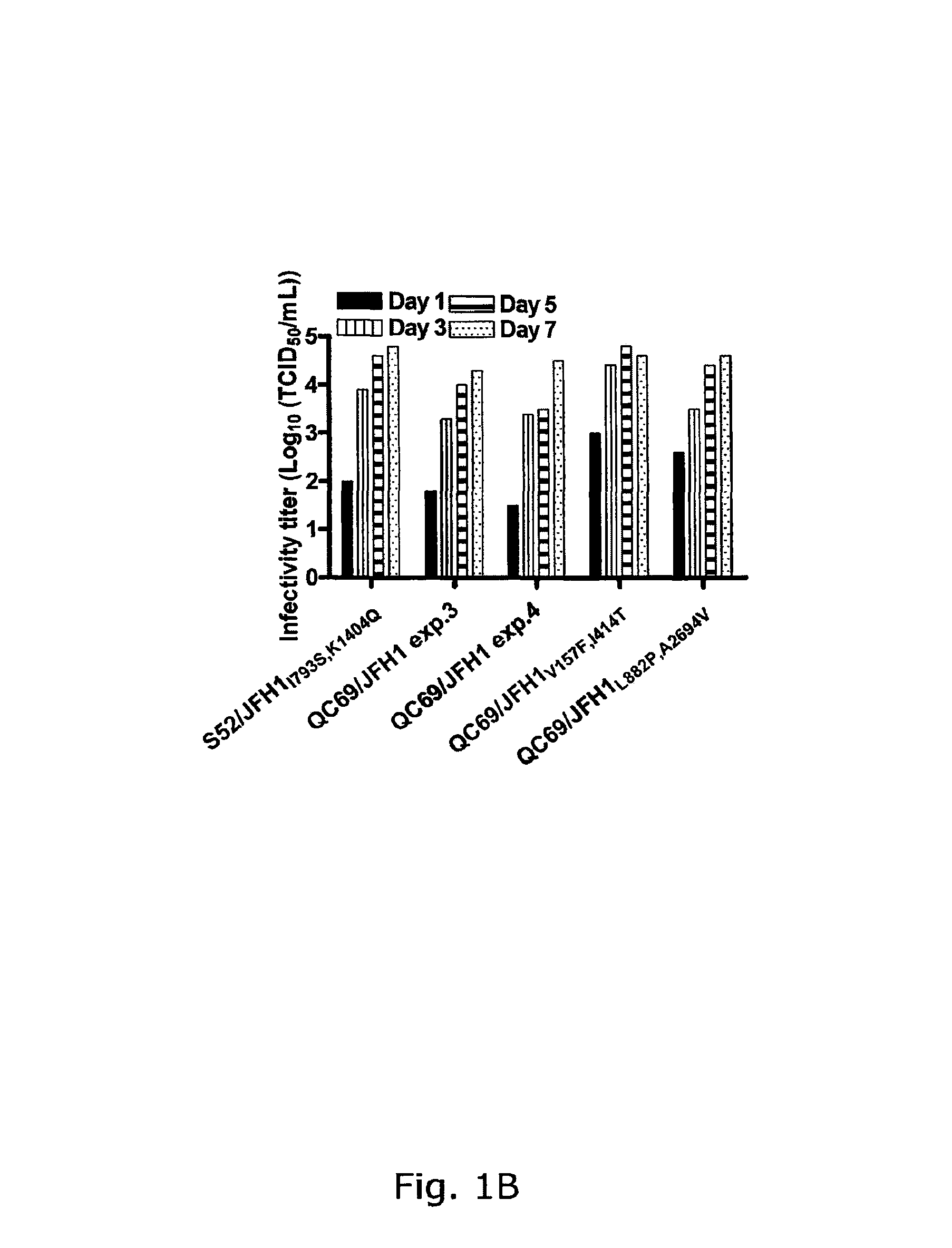
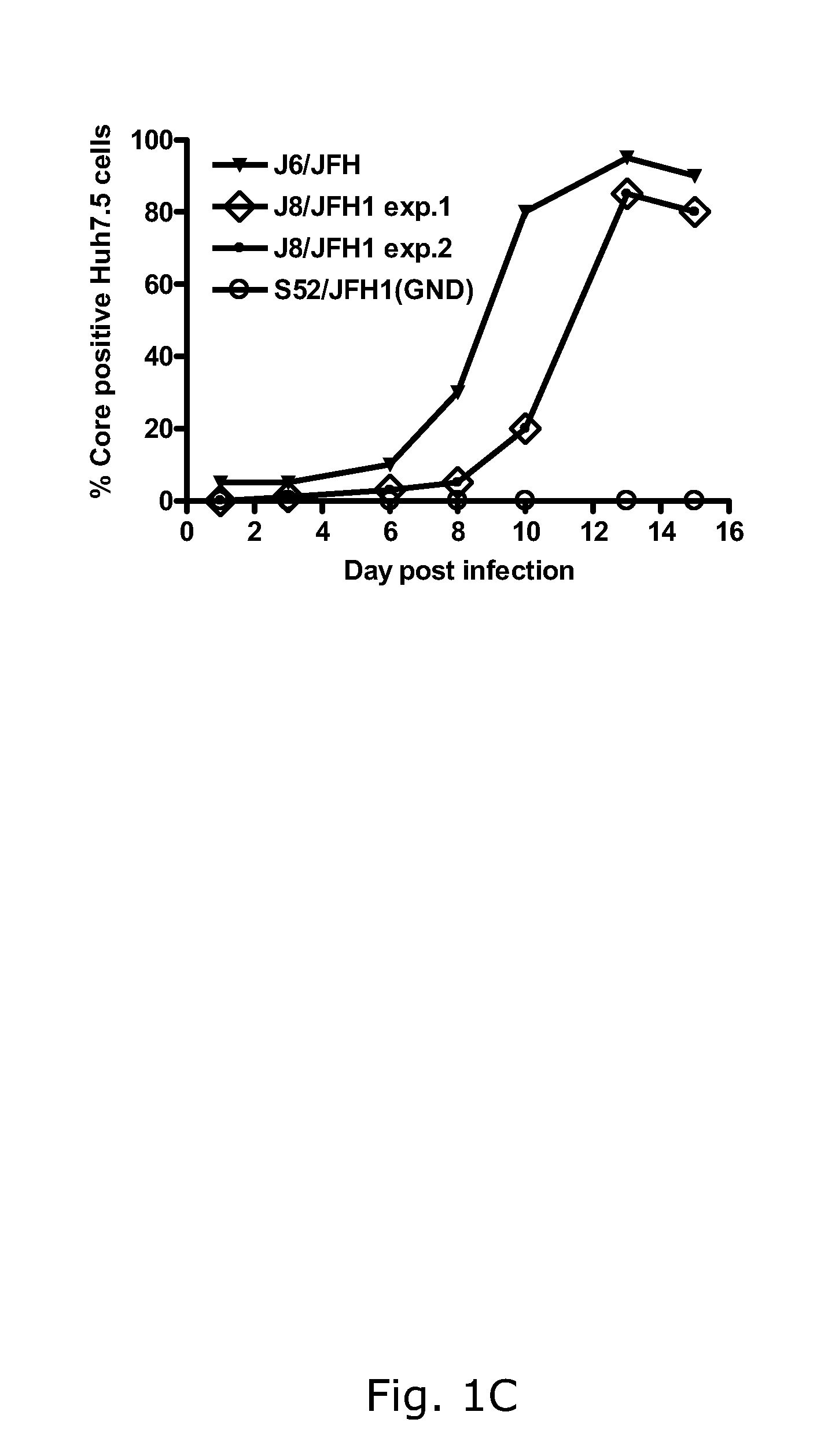
The present inventors have developed a culture system for genotype 3a, which has a high prevalence worldwide. Since intergenotypic recombinant genomes exploiting the replication characteristics of JFH1 will be a valuable tool for the genotype specific study of the replaced genes and related therapeutics, the present inventors constructed a genotype 3a / 2a (S52 / JFH1) recombinant containing the structural genes (Core, E1, E2), p7 and NS2 of strain S52 and characterized it in Huh7.5 cells. S52 / JFH1 and J6 / JFH viruses passaged in cell culture had comparable growth kinetics and yielded similar peak HCV RNA titers and infectivity titers. Direct genome sequencing of cell culture derived S52 / JFH1 viruses identified putative adaptive mutations in Core, E2, p7, NS3 and NS5A; clonal analysis revealed, that all genomes analyzed exhibited different combinations of these mutations. Finally, viruses resulting from transfection with RNA transcripts of five S52 / JFH1 recombinant containing these combinations of putative adaptive mutations performed as efficiently as J6 / JFH viruses in Huh7.5 15 cells and were all genetically stable after viral passage. In conclusion, the present inventors have developed a robust and genetically stable cell culture system for HCV genotype 3a.
Owner:HVIDOVRE HOSPITAL
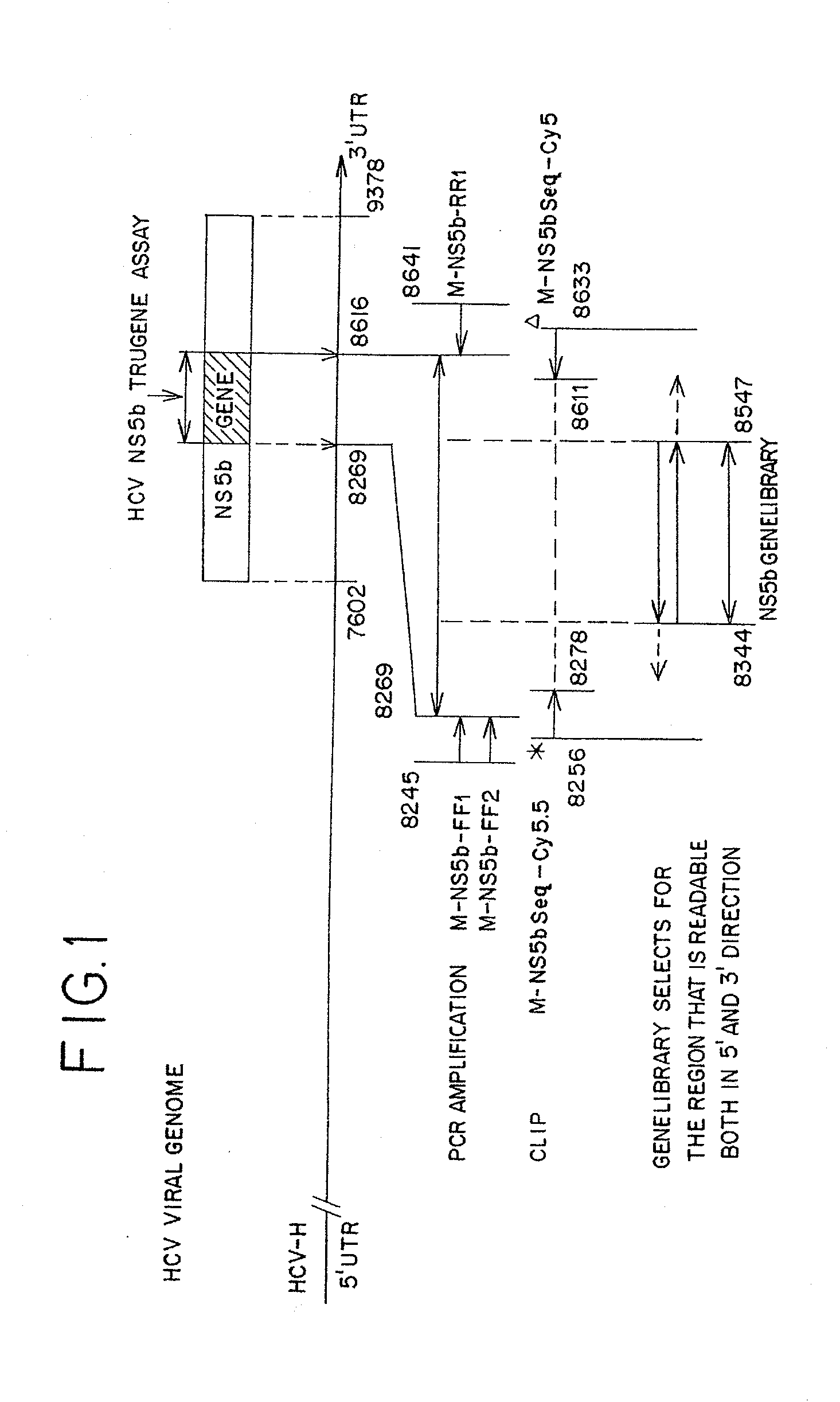
3′terminal sequence of hepatitis C virus genome and diagnostic and therapeutic uses thereof
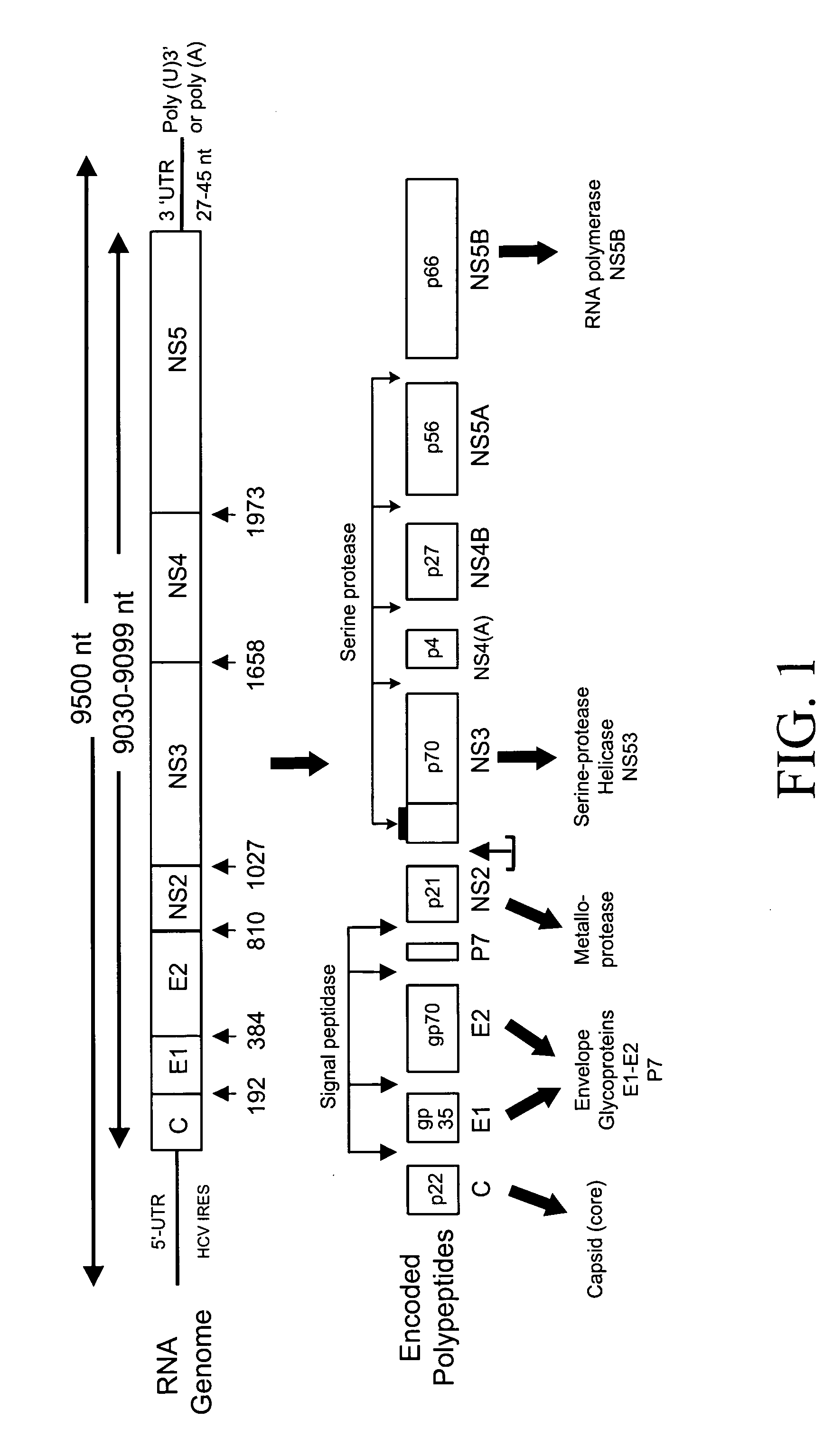
InactiveUS6943246B2Effective strikeAlter adverse consequenceOrganic active ingredientsSsRNA viruses positive-senseHCV GenotypingVaccination
The invention relates to the discovery of a novel RNA sequence at the 3′ terminal sequence of hepatitis C virus (HCV) genome RNA. Included in the invention are the 3′ sequence, its complement, and their use for nucleic-acid based diagnostics and for developing and evaluating novel anti-HCV therapies. This sequence element, which is conserved among HCV genotypes, is likely to be essential for viral replication, and required for construction of full-length HCV cDNA clones capable of yielding infectious RNA, progeny virus or replication-competent HCV replicons. Such functional clones are useful tools for evaluation of therapeutic approaches and as substrates for developing candidate attenuated or inactivated HCV derivatives for vaccination against HCV.
Owner:WASHINGTON UNIV IN SAINT LOUIS
HCV replicons containing NS5B from genotype 2B
The present invention features methods for enhancing the ability of a genotype 2b NS5B sequence to function in a replicon, for producing replicons containing a functional genotype 2b NS5B, and for using replicons to measure the ability of a compound to affect HCV replication that is sustained with the genotype 2b polymerase. Also featured is a genotype 1b NS4B adaptive mutation. The ability to produce replicons containing a functional genotype 2b NS5B is illustrated by the production of chimeric replicons based on HCV genotype 1b where substantially all the NS5B sequence is replaced with a genotype 2b NS5B.
Owner:MERCK SHARP & DOHME CORP +1
HCV genotyping DNA microarray chip
InactiveCN101660002AMicrobiological testing/measurementDNA/RNA fragmentationDNA Microarray ChipHCV Genotyping
The invention relates to a DNA microarray chip, in particular to a hepatitis C virus (HCV) genotyping DNA microarray chip. A solid-phase carrier substrate is used, and an oligonucleotide probe is designed in view of different genotypes of HCV so as to prepare the DNA microarray chip; and when matched with a PCR primer and other components, the DNA microarray chip can quickly and accurately classify the types of the hepatitis C virus in a blood sample.
Owner:DAAN GENE CO LTD
Efficient cell culture system for hepatitis c virus genotype 6a
InactiveUS20110059512A1Raise the potentialSsRNA viruses positive-senseSugar derivativesSequence analysisHCV Genotyping
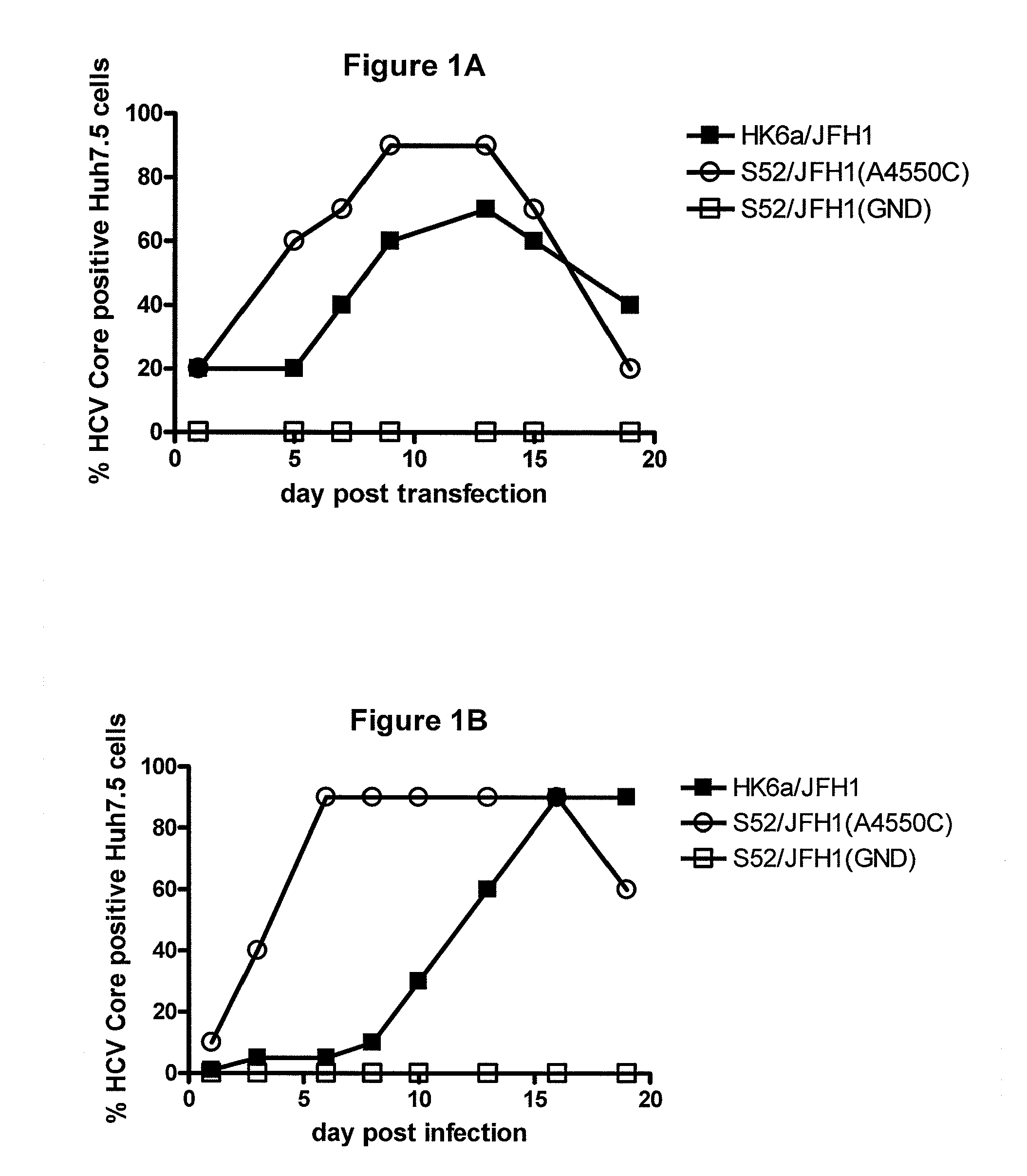
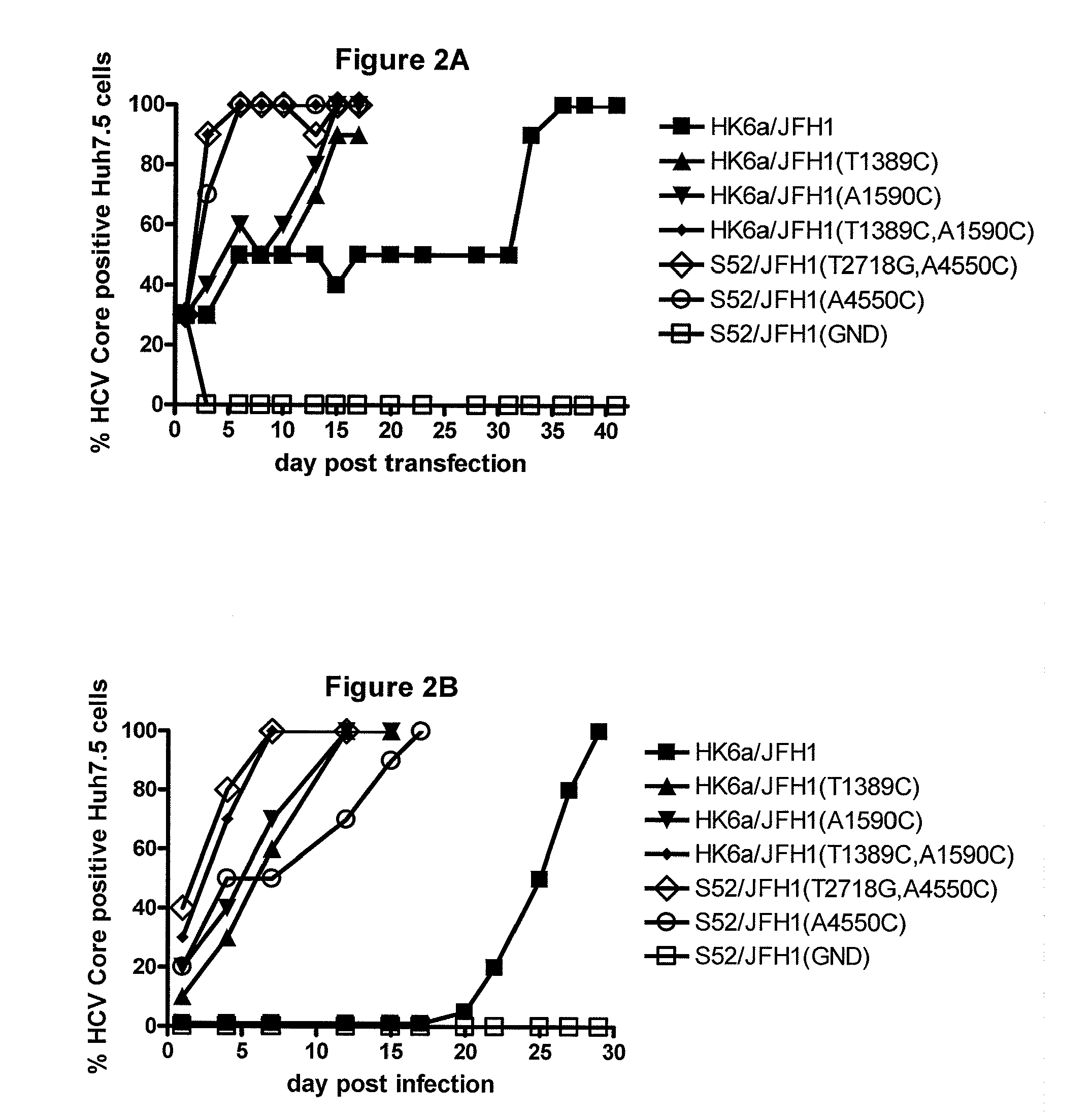
The present inventors developed hepatitis C virus 6a / 2a intergenotypic recombinants in which the JFH1 structural genes (Core, E1 and E2), p7 and the complete NS2 were replaced by the corresponding genes of the genotype 6a reference strain HK6a. Sequence analysis of recovered 6a / 2a recombinants from 2 transfection experiments and subsequent reverse genetic studies revealed adaptive mutations in E1 and E2. Conclusion: The developed 6a / 2a viruses provide a robust in vitro tool for research in HCV genotype 6, including vaccine studies and functional analyses.
Owner:HVIDOVRE HOSPITAL
EFFICIENT CELL CULTURE SYSTEM FOR HEPATITIS C VIRUS GENOTYPE 7a
InactiveUS20110294195A1High infectivity titerEfficient growth processSsRNA viruses positive-senseBacteriaSequence analysisHepatitis C virus genotype
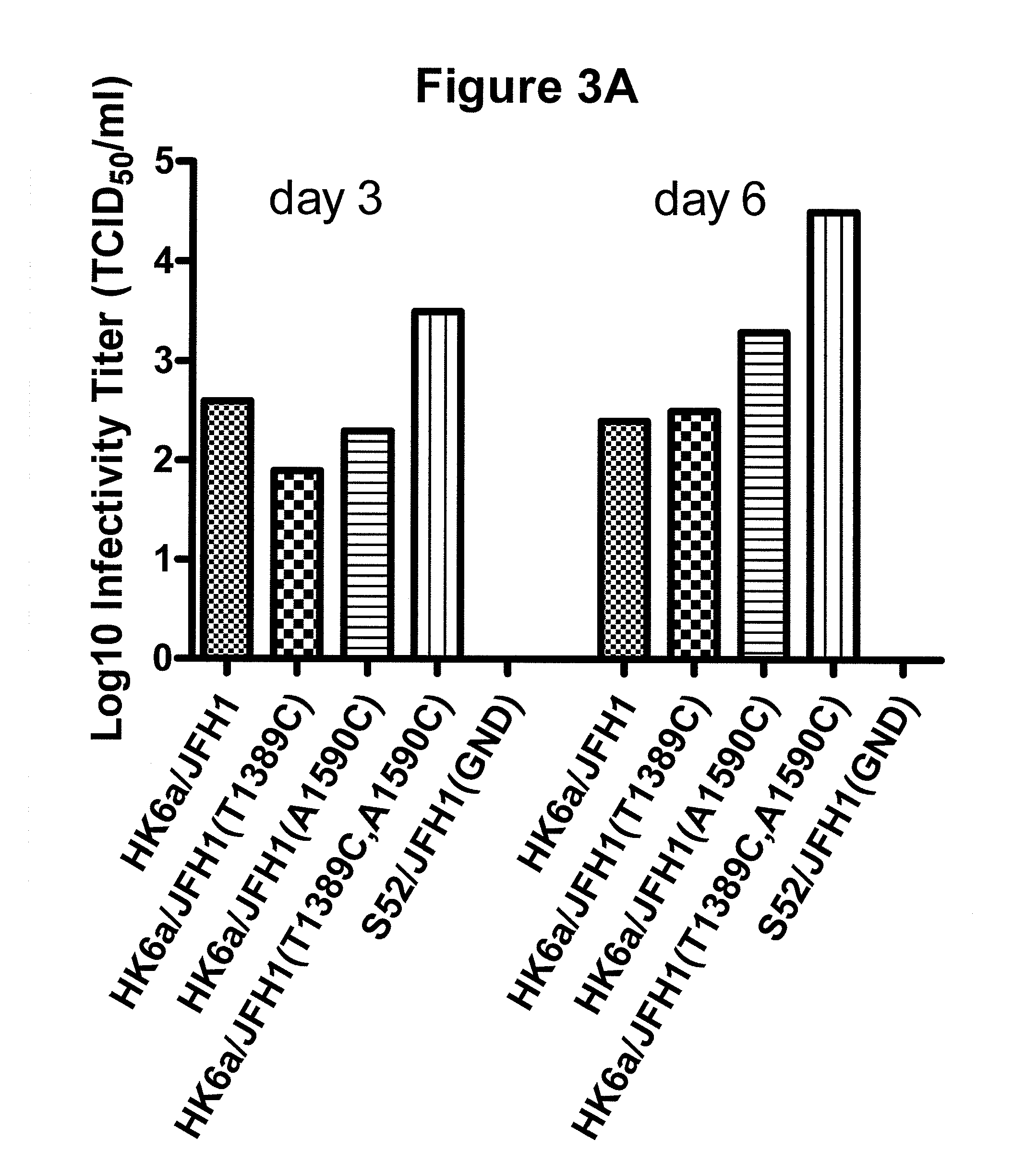
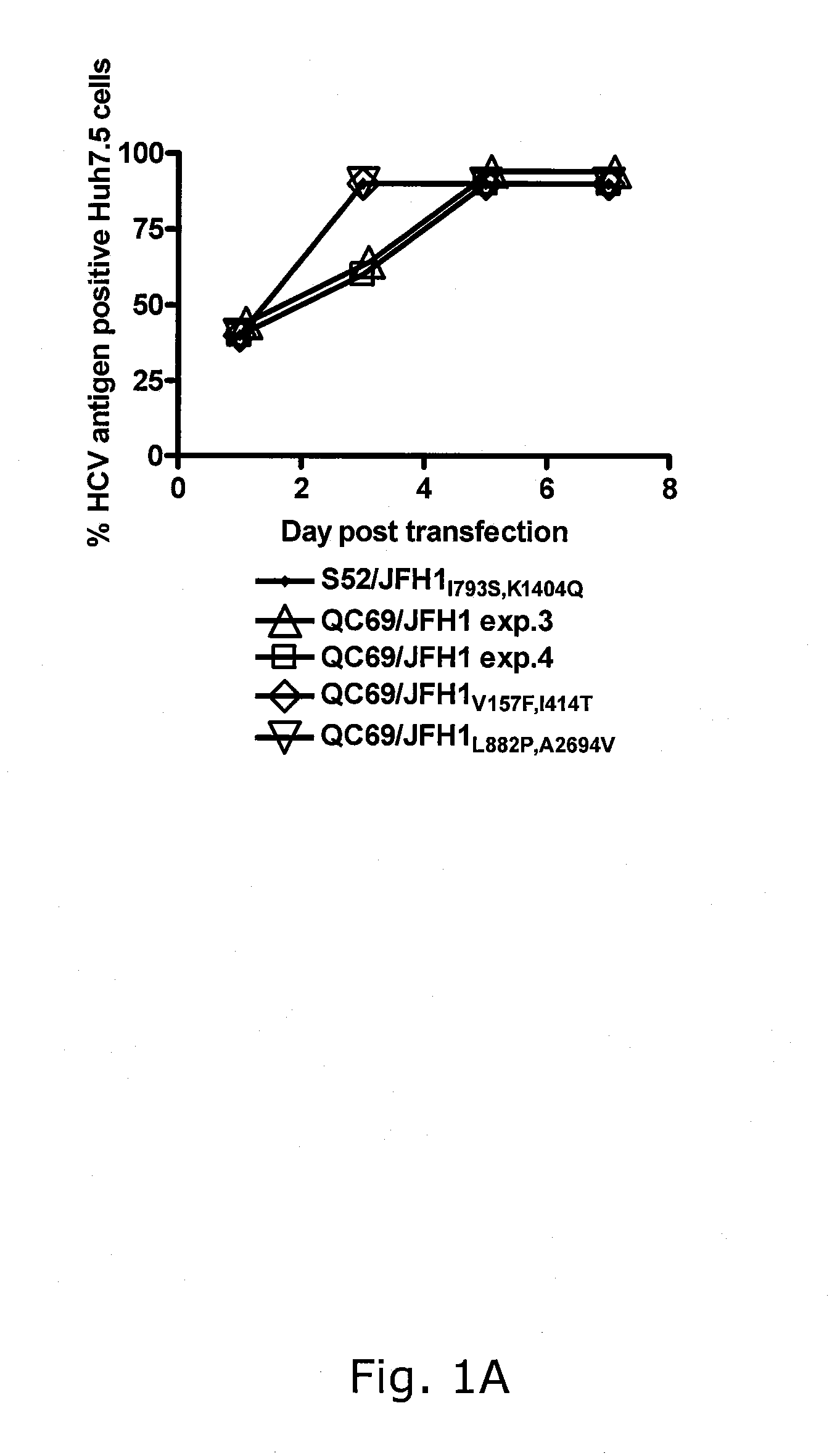
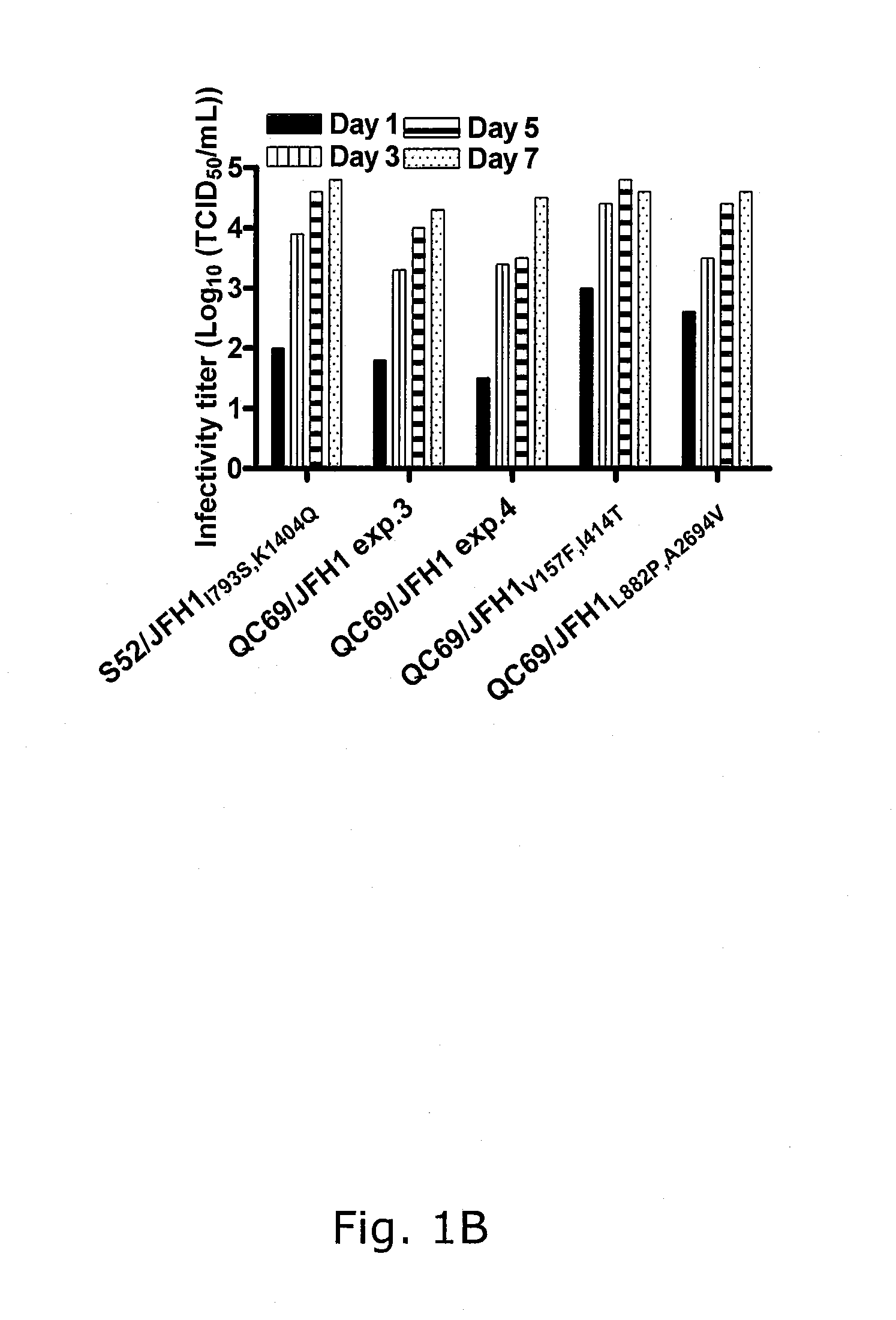
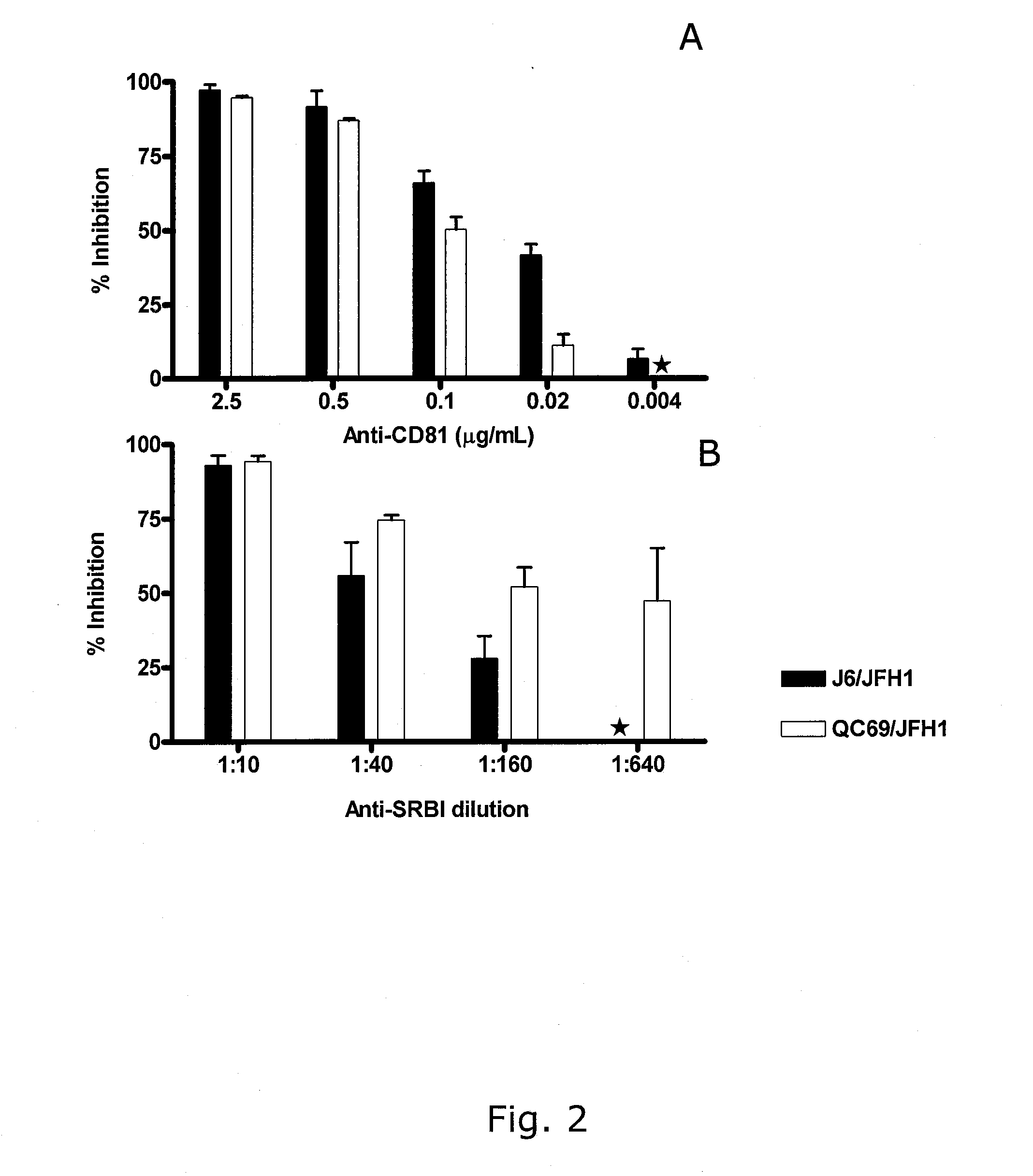
Genotype 7a has been identified recently, thus not much is known about the biology of this new, major HCV genotype. The present inventors developed hepatitis C virus 7a / 2a intergenotypic recombinants in which the JFH1 structural genes (Core, E1 and E2), p7 and the complete NS2 were replaced by the corresponding genes of the genotype 7a strain QC69 and characterized them in Huh7.5 cells. Sequence analysis of 7a / JFH1 recombinants recovered after viral passage in Huh7.5 cells following 4 independent transfection experiments revealed adaptive mutations in Core, E2, NS2, NS5A and NS5B. In reverse genetic studies the importance of these mutations for improved growth kinetics was shown. Adapted 7a / JFH1 viruses showed growth kinetics, infectivity and RNA titers comparable to a previously developed 3a / JFH1 reference virus. Conclusion: The developed 7a / JFH1 viruses provide a robust in vitro tool for research in HCV genotype 7, including vaccine studies and functional analyses.
Owner:HVIDOVRE HOSPITAL
Efficient cell culture system for hepatitis c virus genotype 5a
InactiveUS20110021611A1Organic active ingredientsSsRNA viruses positive-senseSequence analysisSerial passage
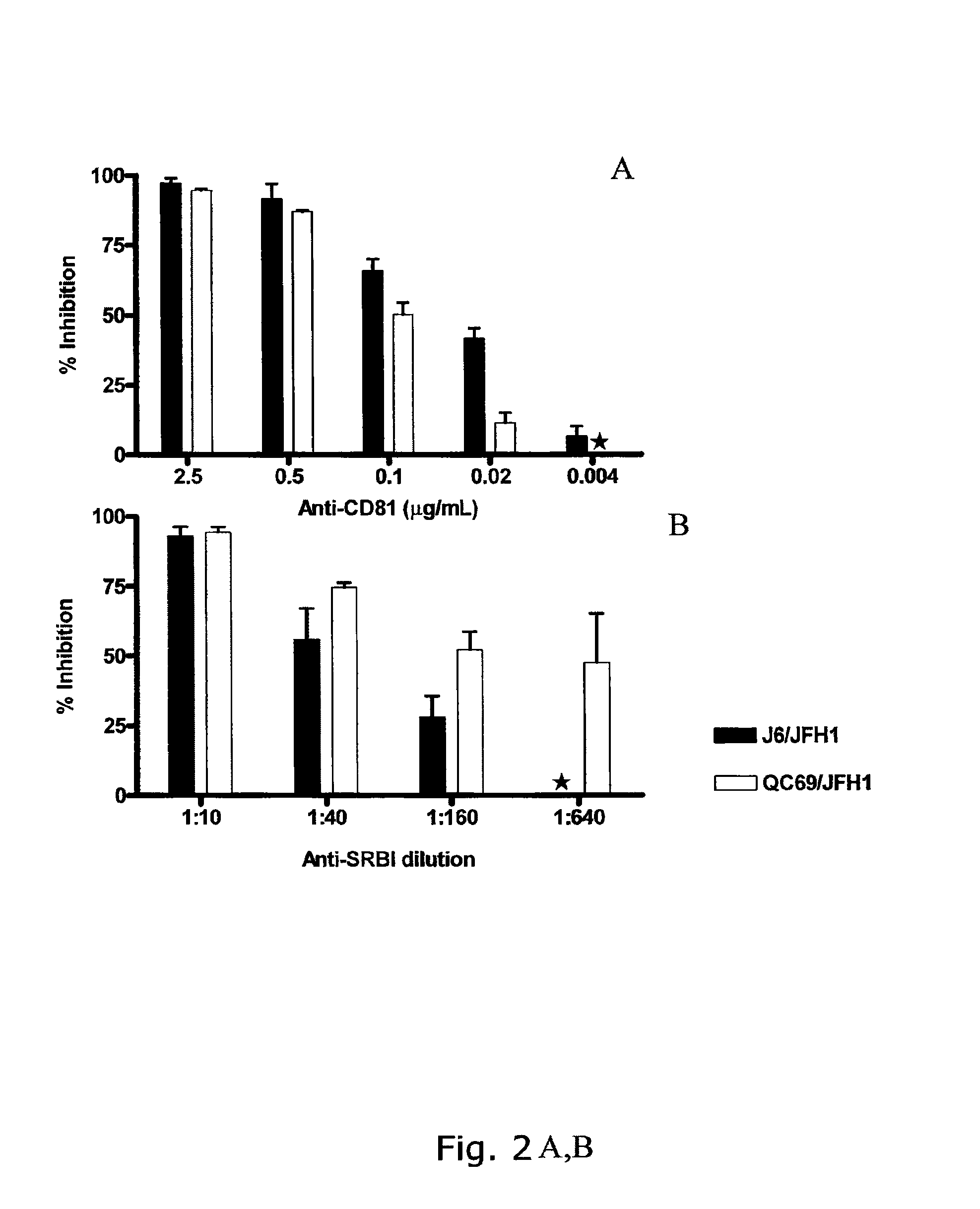
The present inventors developed 5a / 2a intergenotypic recombinants in which the JFH1 structural genes (Core, E1 and E2), p7 and all of or part of NS2 were replaced by the corresponding genes of the genotype 5a reference strain SA13. Compared to the J6 / JFH control virus, after transfection of in vitro transcripts in Huh7.5 cells, production of infectious viruses was delayed. However, in subsequent viral passages efficient spread of infection and HCV RNA titers as high as for J6 / JFH were obtained. Infectivity titers were at all time points analyzed comparable to J6 / JFH control virus. Sequence analysis of recovered 5a / 2a recombinants from 2 serial passages and subsequent reverse genetic studies revealed adaptive mutations in p7, NS2 and / or NS3. Infectivity of the 5a / 2a viruses was CD81 and SR-BI dependant, and the recombinant viruses could be neutralized by chronic phase sera from patients infected with genotype 5a. Conclusion: The developed 5a / 2a viruses provide a robust in vitro tool for research in HCV genotype 5, including vaccine studies and functional analyses of an increasingly important genotype in South Africa and Europe.
Owner:HVIDOVRE HOSPITAL
Efficient cell culture system for hepatitis C virus genotype 6A
The present inventors developed hepatitis C virus 6a / 2a intergenotypic recombinants in which the JFH1 structural genes (Core, E1 and E2), p7 and the complete NS2 were replaced by the corresponding genes of the genotype 6a reference strain HK6a. Sequence analysis of recovered 6a / 2a recombinants from 2 transfection experiments and subsequent reverse genetic studies revealed adaptive mutations in E1 and E2. Conclusion: The developed 6a / 2a viruses provide a robust in vitro tool for research in HCV genotype 6, including vaccine studies and functional analysis.
Owner:HVIDOVRE HOSPITAL
A kit for genotyping hepatitis C virus
ActiveCN102286644AHigh detection sensitivityImprove accuracyMicrobiological testing/measurementMicroorganism based processesForward primerHCV Genotyping
The invention relates to a kit for genotyping hepatitis C virus (HCV), which comprises HCVNS5B gene amplification primers, HCVNCR conserved region gene amplification primers, an HCVRNA extraction reagent, a negative control, a positive control, a cDNA (complementary deoxyribonucleic acid) synthetic reagent, a PCR (polymerase chain reaction) solution and a PCR sequencing reagent, wherein the sequence of the HCVNS5B gene forward primer is disclosed as SEQ ID NO.1, the sequence of the HCVNS5B gene reverse primer is disclosed as SEQ ID NO.2, the sequence of the HCVNCR conserved region gene forward primer is disclosed as SEQ ID NO.3, and the sequence of the HCVNCR conserved region gene forward primer is disclosed as SEQ ID NO.4. The kit provided by the invention has the advantages of high detection sensitivity and good specificity, and can be used for detecting all the reported HCV genotypes (subtypes) at high speed (within 12-14 hours).
Owner:HUAXIN SCI & TECH PANYU CITY
Oligonucleotides chip composition for analyzing HCV gene type and its detecting method
InactiveCN1535319AMicrobiological testing/measurementRecombinant DNA-technologyHCV GenotypingOligonucleotide chip
The invention discloses a method for analyzing HCV genotype by extracting RNA from plasma and serum, carrying out RT-PCR of hepatitis C virus (HCV) and reacting it on an oligonucleotide chip. The present invention also provides a simple and accurate method for assaying 4 human HCV genotypes on one slide.
Owner:株式会社生物核心
Efficient cell culture system for hepatitis C virus genotype 7a
Owner:HVIDOVRE HOSPITAL
Adaptive mutations allow establishment of JFH1-based cell culture systems for hepatitis C virus genotype 4A
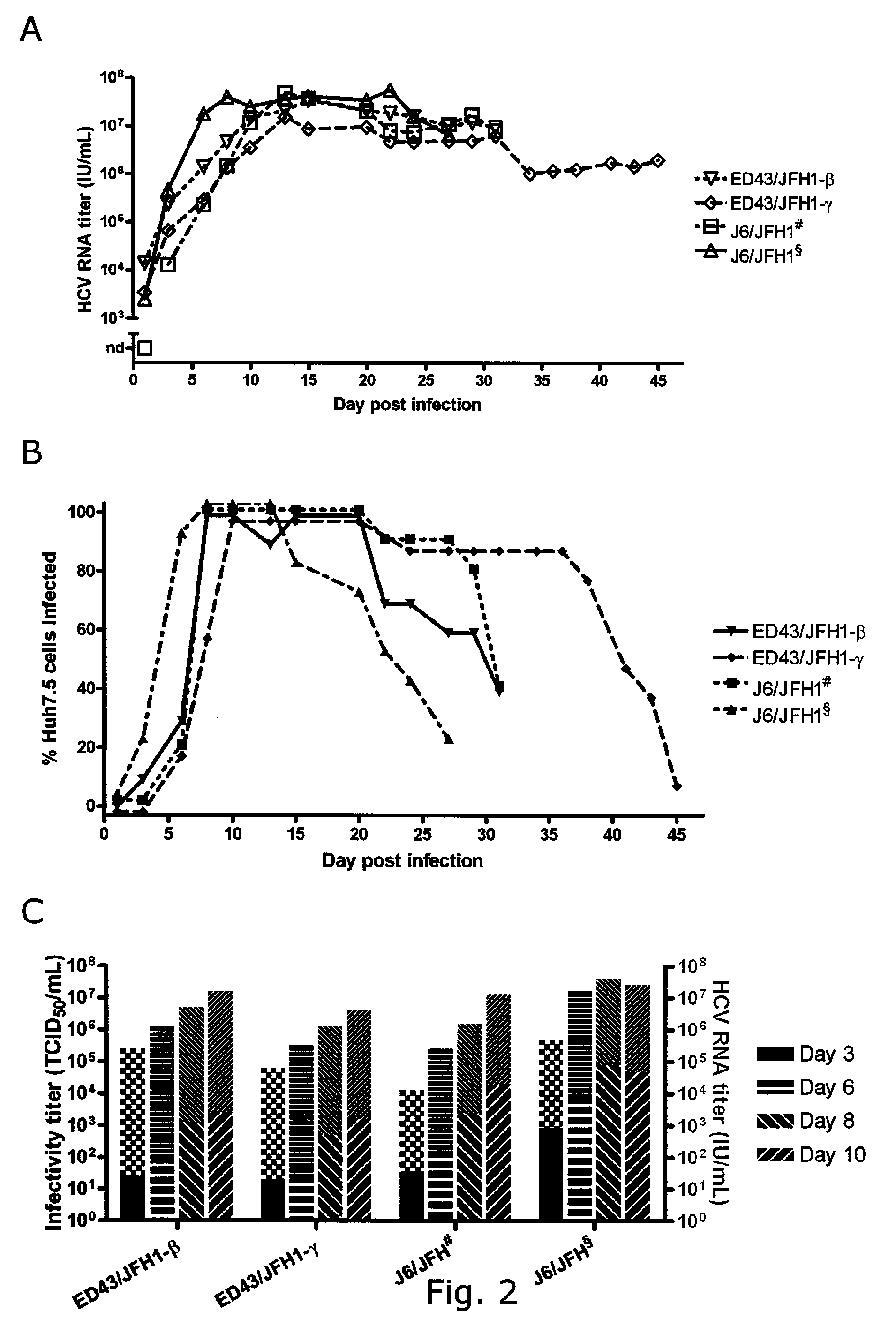
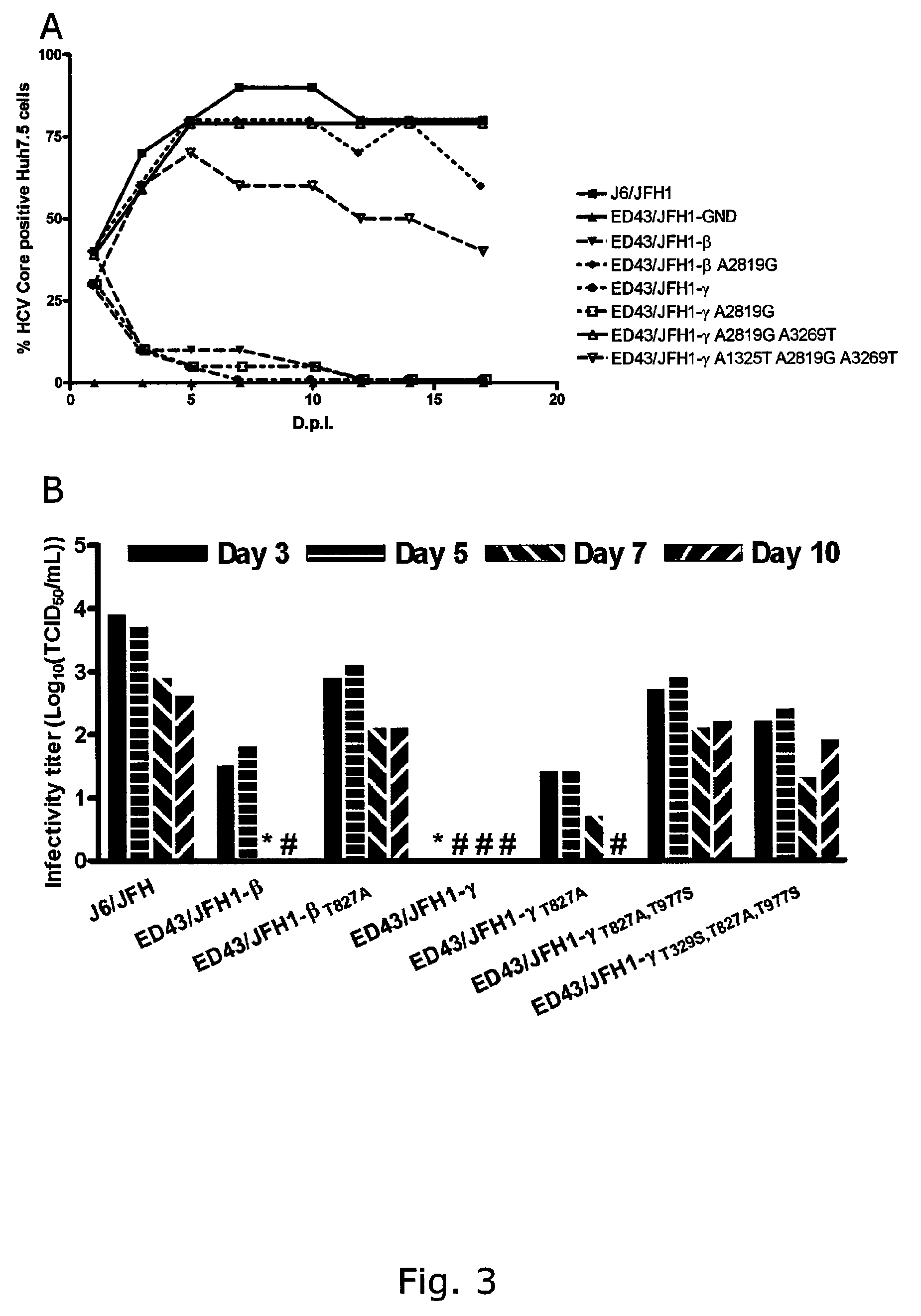
The present inventors developed three 4a / 2a intergenotypic recombinants in which the JFH1 structural genes (Core, E1 and E2), p7 and all of or part of NS2 were replaced by the corresponding genes of the genotype 4a reference strain ED43. The 4a / 2a junction in NS2 was placed after the first transmembrane domain (α), in the cytoplasmic part (β) or at the NS2 / NS3 cleavage site (y). Following transfection of Huh7.5 cells with RNA transcripts, infectious viruses were produced in the ED43 / JFH1-β and -y cultures only. Compared to the 2a control virus, production of infectious viruses was significantly delayed. However, in subsequent passages efficient spread of infection and high HCV RNA titers were obtained. Infectivity titers were approximately 10-fold lower than for the 2a control virus. Sequence analysis of recovered 4a / 2a recombinants from 3 serial passages and subsequent reverse genetic studies revealed a vital dependence on a mutation in the NS2 4a part. ED43 / JFH1-γ further depended on a second NS2 mutation. Infectivity of the 4a / 2a viruses was CD81 dependent. Conclusion: The developed 4a / 2a viruses provide a robust in vitro tool for research in HCV genotype 4, including vaccine studies and functional analyses of an increasingly important genotype in the Middle East and Europe.
Owner:HVIDOVRE HOSPITAL
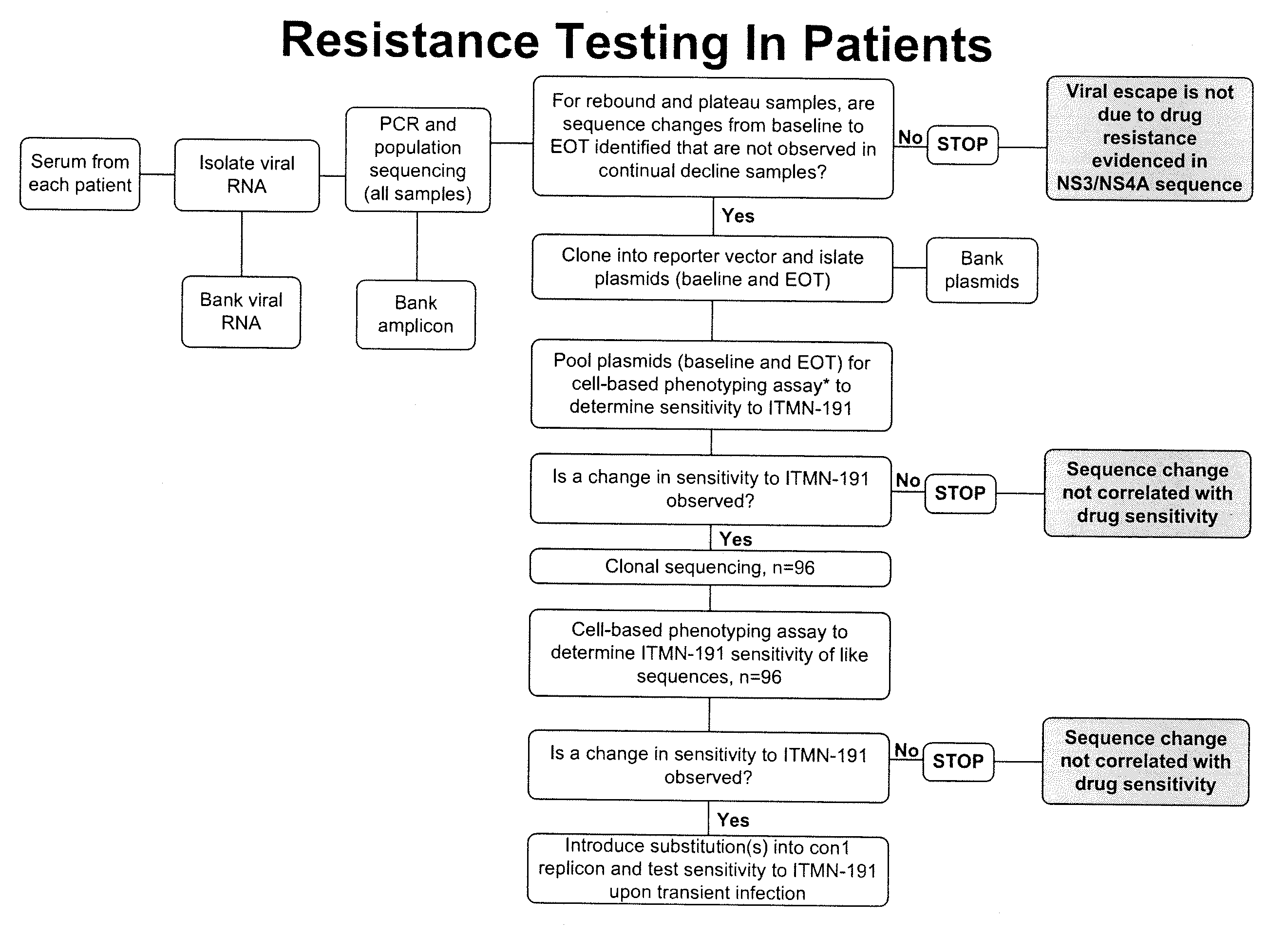
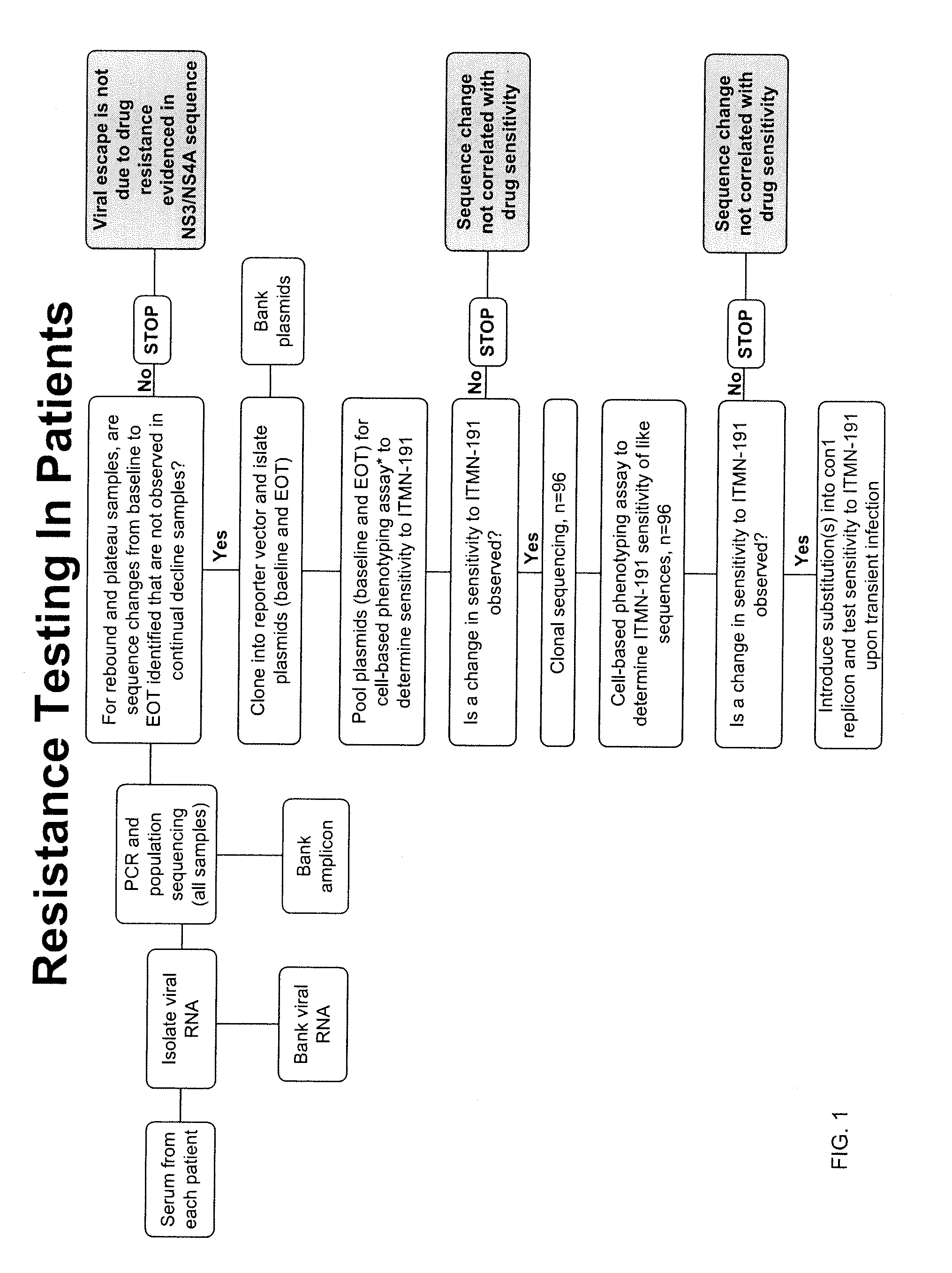
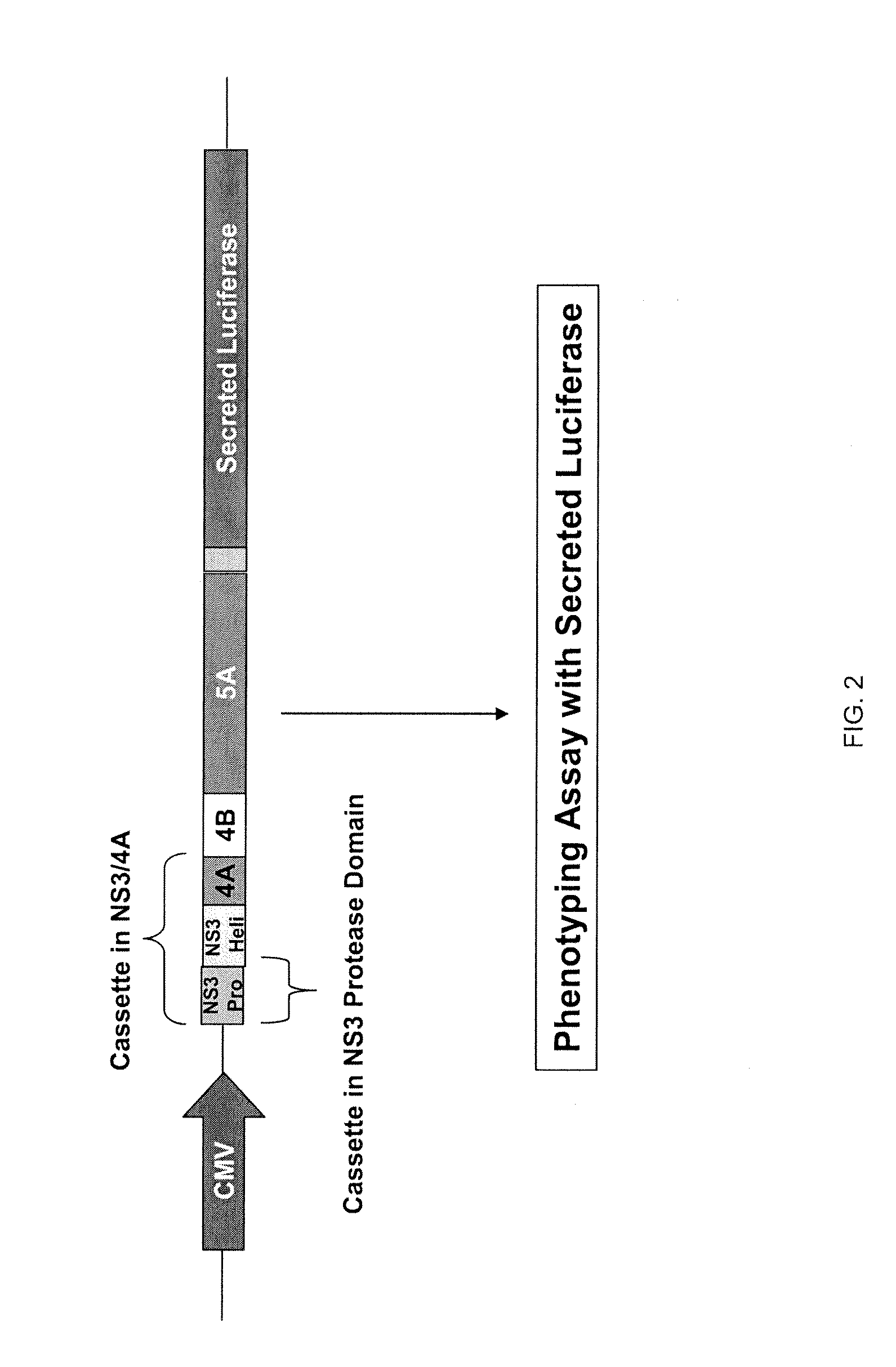
Hcv genotyping and phenotyping
InactiveUS20090220943A1Minimize and reduce non-specific background noise signalSugar derivativesMicrobiological testing/measurementHCV GenotypingAntiviral drug
The present invention includes methods of genotyping and phenotyping HCV. In one embodiment, the methods of the invention can be used to determine whether a HCV isolate is resistant to an antiviral drug. The invention also includes primers for amplifying a HCV NS3 region and kits.
Owner:INTERMUNE INC
Hcv ns3-ns4a protease inhibition
InactiveCN101072575AInhibition of replicationReduce riskDipeptide ingredientsTetrapeptide ingredientsHCV GenotypingProteinase activity
The present invention relates to inhibiting the activity of non-genotype 1 hepatitis C virus (HCV) NS3-NS4A protease activity. More particularly, the invention relates to inhibiting the activity of the protease from HCV genotype-2 or HCV genotype-3. The methods of the invention emply inhibitors that act by interfering with the life cycle of the HCV and are also useful as antiviral agents. The invention further relates to compositions comprising such compounds either for ex vivo use or for administration to a patient suffering from genotype-2 or genotype-3 HCV infection. The invention also relates to methods of treating an HCV infection in a patient by administering a composition comprising a compound of this invention.
Owner:VERTEX PHARMA INC
Methods and Reagents for Genotyping HCV
ActiveUS20100196876A1SsRNA viruses positive-senseMicrobiological testing/measurementHCV GenotypingNucleotide
The present invention is directed to methods and reagents for determining the genotype of a hepatitis C virus (HCV) species present in a test sample. The invention more particularly relates to mixtures of degenerate amplification and sequencing primers, and methods of using such primers, that are complementary to a plurality of HCV species, and are capable of generating nucleotide sequence information for a region of NS5B of HCV that is, for each species, indicative of the type and / or subtype, of the species present in the sample.
Owner:SIEMENS HEALTHCARE DIAGNOSTICS INC
Efficient cell culture system for hepatitis C virus genotype 2B
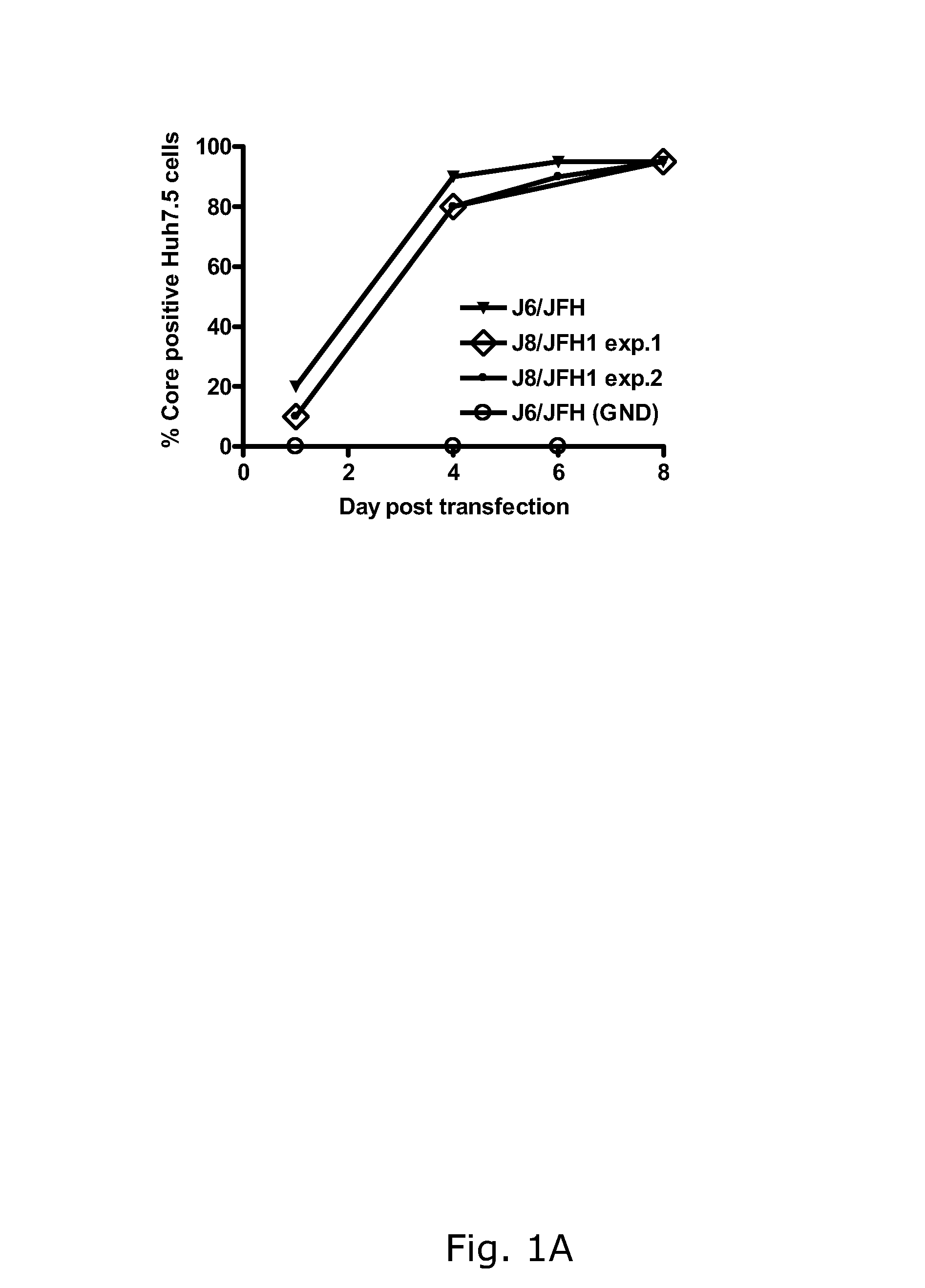
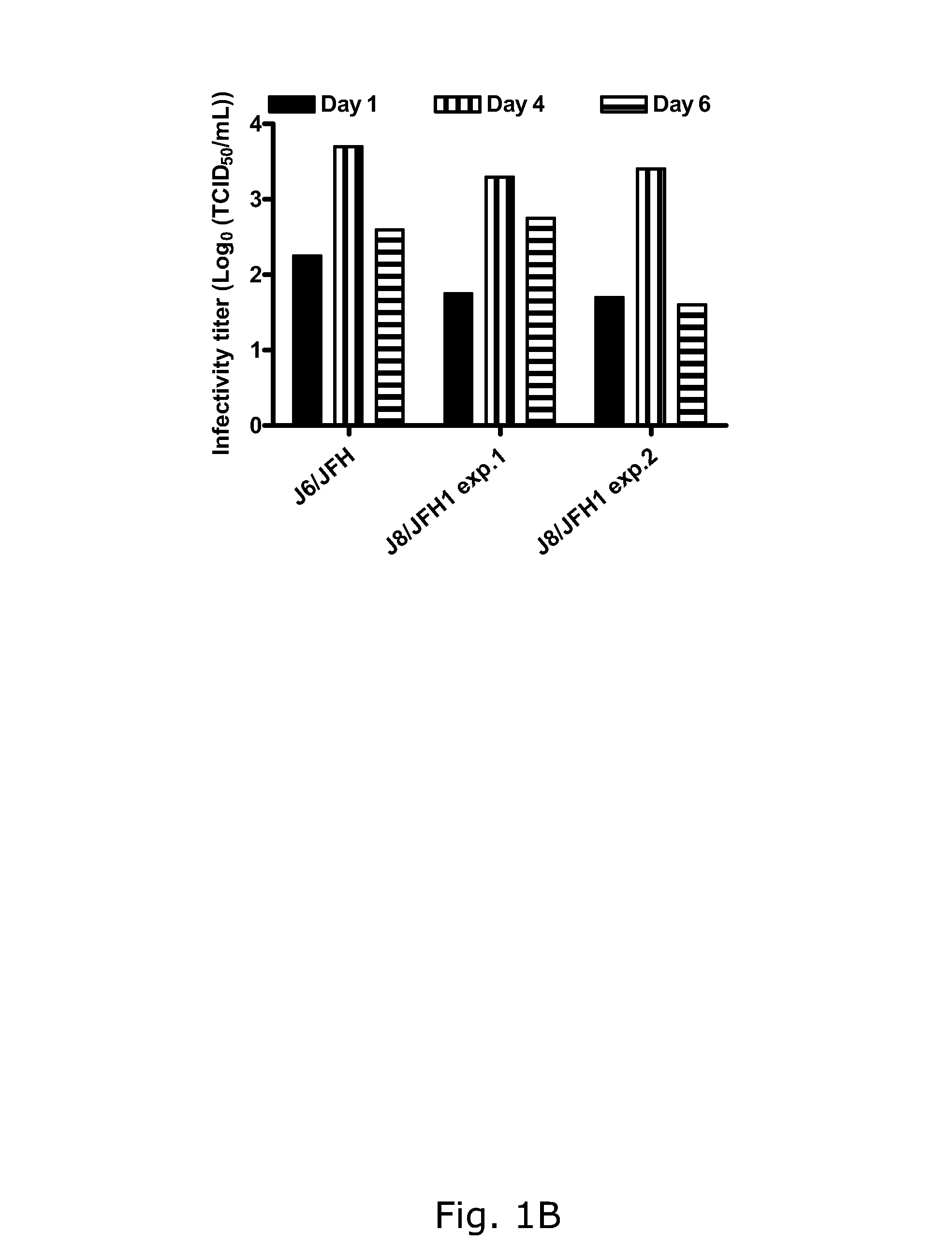
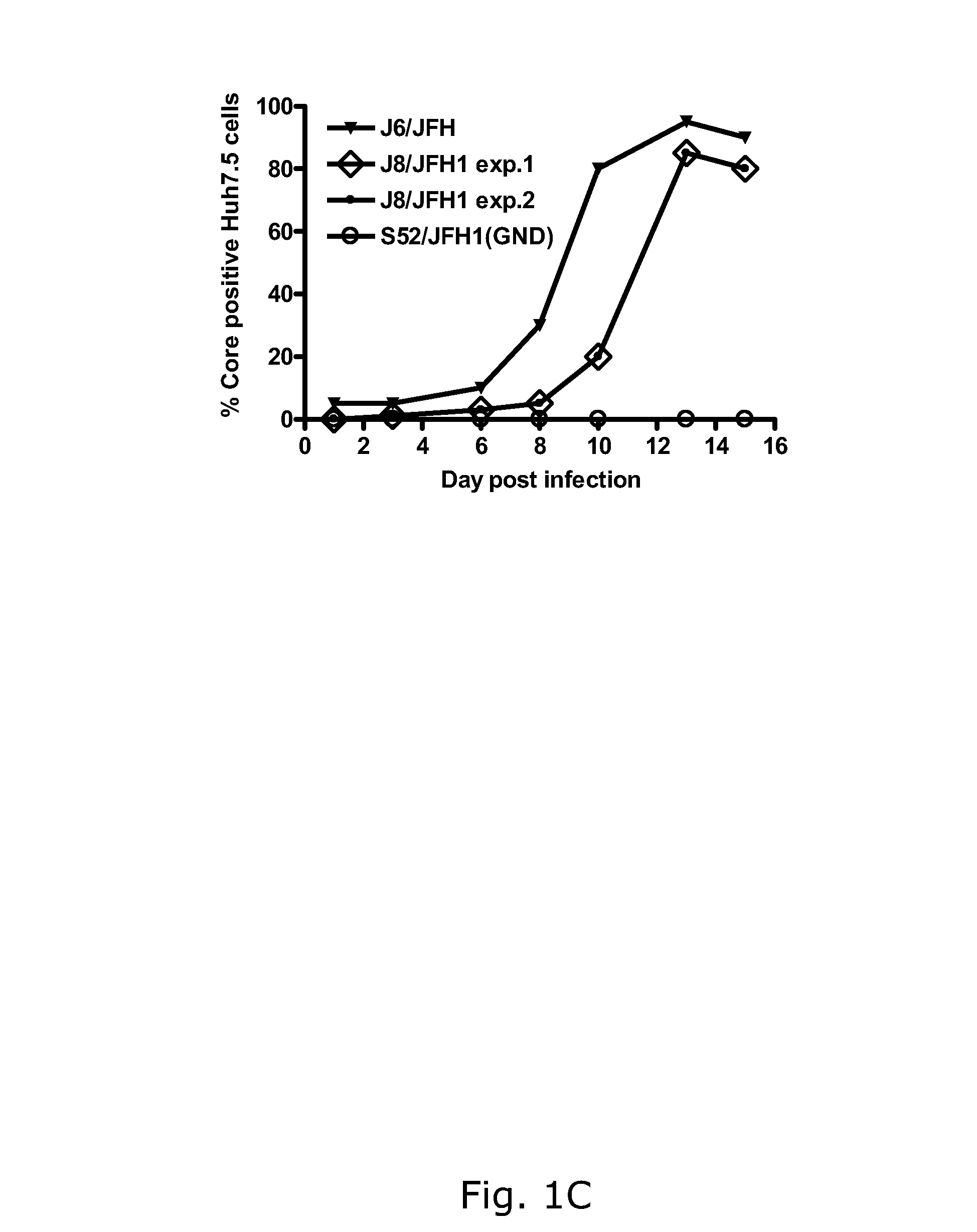
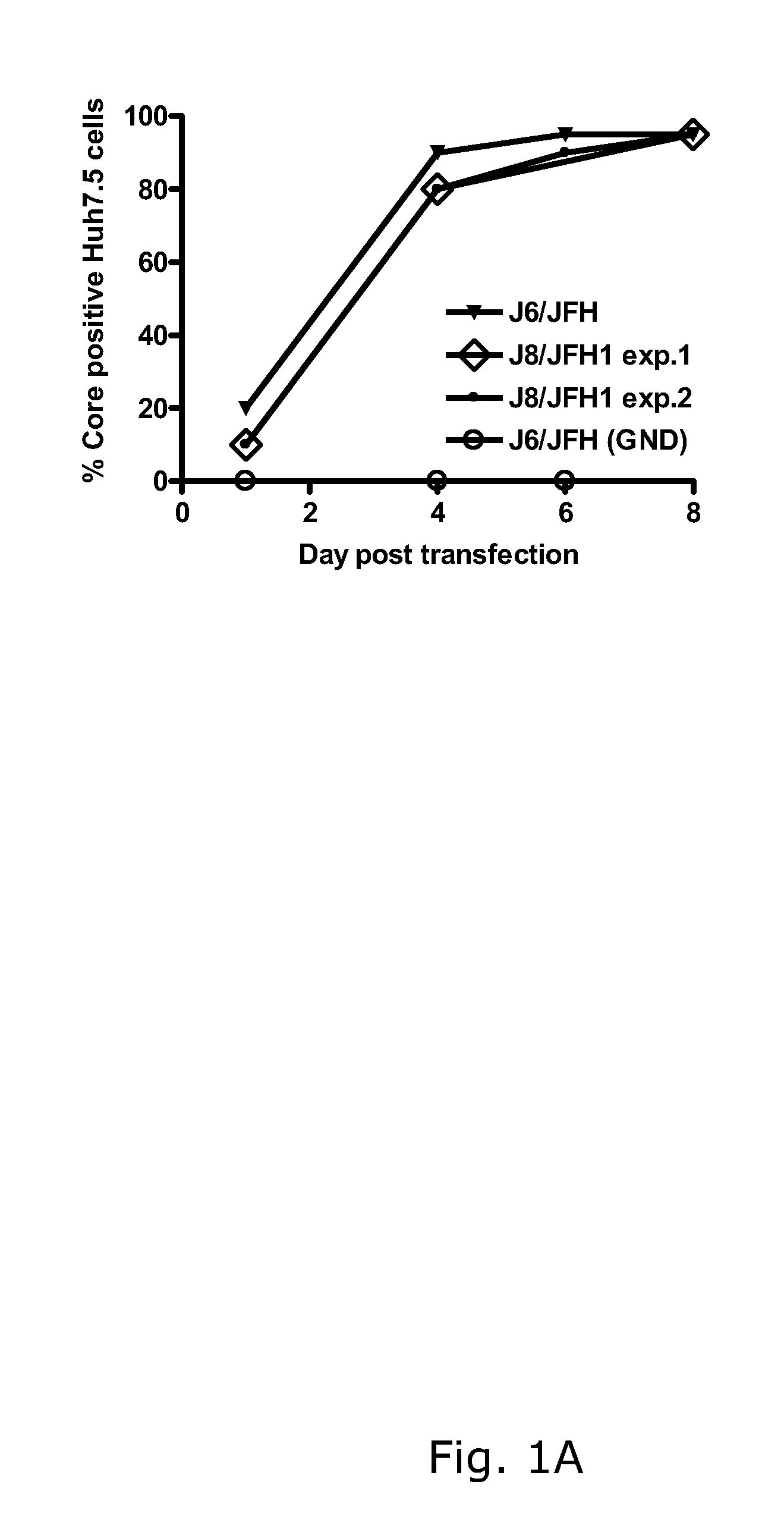
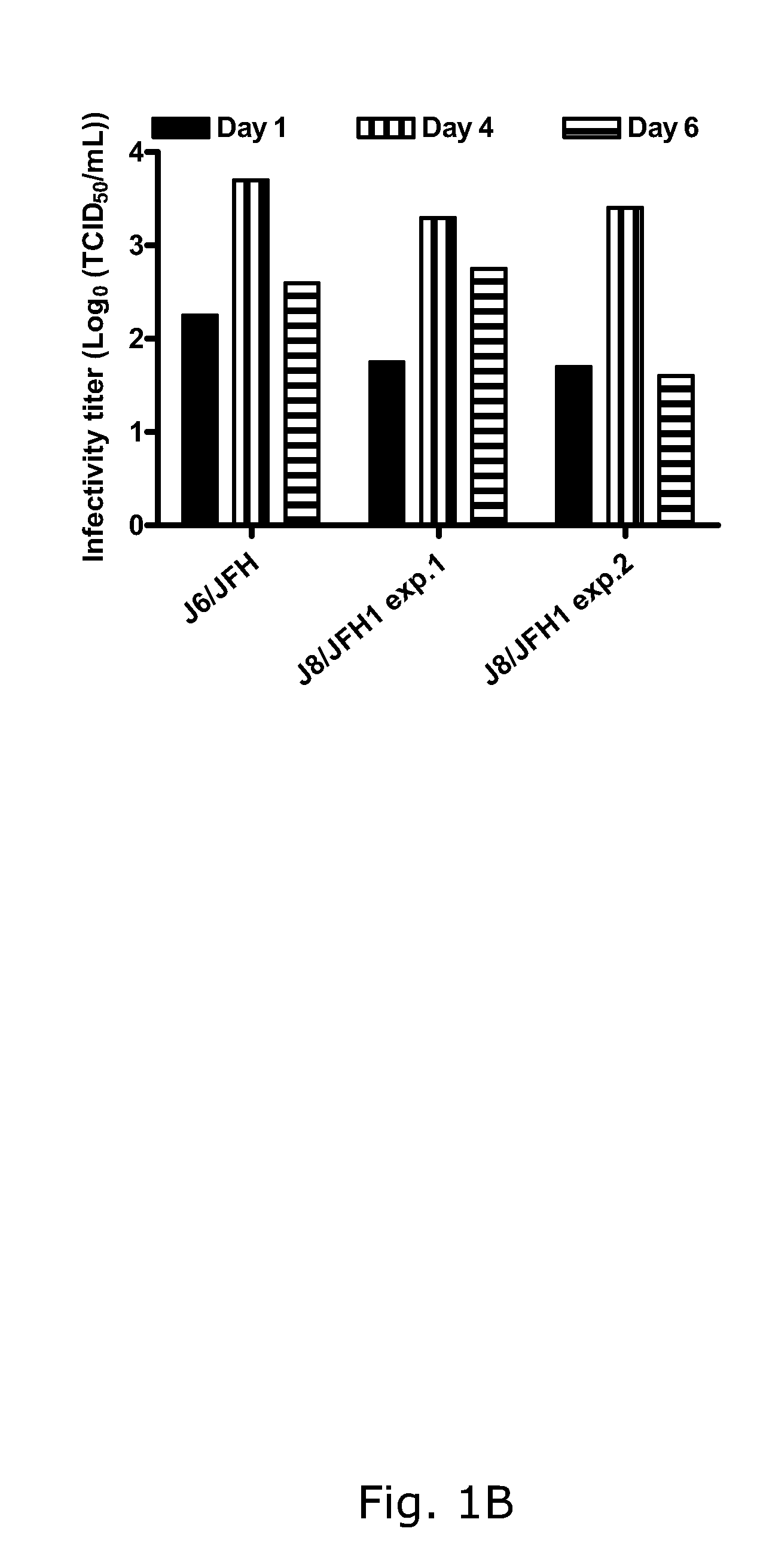
The present inventors developed hepatitis C virus 2b / 2a intergenotypic recombinants in which the JFH1 structural genes (Core, E1 and E2), p7 and the complete NS2 were replaced by the corresponding genes of the genotype 2b reference strain J8. Sequence analysis of recovered 2b / 2a recombinants from 2 transfection experiments revealed that 2b / 2a was genetically stable. Conclusion: The developed 2b / 2a viruses provide a robust in vitro tool for research in HCV genotype 2b, including vaccine studies and functional analysis.
Owner:HVIDOVRE HOSPITAL
Hepatitis C virus (HCV) genotyping kit
ActiveCN103966363AHigh sensitivityStrong specificityMicrobiological testing/measurementMicroorganism based processesHCV GenotypingHybridization reaction
The invention relates to a disease pathogen gene detecting technology and particularly relates to a hepatitis C virus (HCV) genotyping kit which can be used for auxiliary diagnosis for infection of types 1a, 1b, 2a, 2b, 3a, 3b and 6a of HCV happening in China. According to the invention, a nylon membrane carrier is adopted, an oligonucleotide probe is designed according to the different genotypes of the HCV, a gene chip is prepared, and then, a PCR (Polymerase Chain Reaction) primer and other components are matched, so that the HCV in a blood sample can be rapidly and accurately typed. The HCV genotyping kit provided by the invention has the characteristics of rapidness, sensitivity, accuracy, high flux, simplicity, convenience, high sensitivity and strong specificity, and is capable of detecting various target sequences in hybridization reaction once, convenient to take materials, free of special equipment and low in instrument investment.
Owner:福州泰普生物科学有限公司
Nucleotide sequence and kit for detecting hepatitis C viruses
InactiveCN107653344AStrong specificityImprove the success rate of amplificationMicrobiological testing/measurementDNA/RNA fragmentationHCV GenotypingMolecular diagnostic techniques
The invention discloses a nucleotide sequence and a kit for detecting hepatitis C viruses (HCV), and belongs to the technical field of molecular diagnosis. By a primer and a probe for detecting the HCV, virus RNA of seven HCV genotypes can be specifically amplified and detected. By the kit for detecting the hepatitis C viruses, accurate copy numbers of various subtypes of the HCV can be detected quickly, sensitively and accurately. The nucleotide sequence and the kit have the advantages of simplicity and convenience in operation, high specificity, high sensitivity, low cost, high throughput and the like, can be applied to clinical quick hyper-sensitive detection of HCV virus RNA, and provide reference to clinical diagnosis and treatment of hepatitis C.
Owner:HANGZHOU D A GENETIC ENG
Method for rapidly and sensitively detecting hepatitis C virus (HCV) and genotype identification method
InactiveCN106282404AIncrease success rateSpecific and accurate detectionMicrobiological testing/measurementMicroorganism based processesHCV GenotypingHcv genotype 1
The invention provides oligonucleotides and a testing agent used for rapidly and sensitively detecting, identifying or distinguishing various genotypes and subtypes of hepatitis C viruses (HCVs). In one embodiment, the invention provides universal primers and probes corresponding to all the HCVs and the universal primers and probes can capture universal or common nucleic acids from the HCV ethnic groups. In one embodiment, the primers and the probes can capture the nucleic acids from rarely seen, unclassified or unknown HCV genotypes, subtypes or strains. In another embodiment, the invention provides probes capable of clearly detecting or identifying the HCV genotypes 1-6 or distinguishing the genotypes. In one embodiment, the invention provides primers and probes capable of clearly detecting or identifying the HCV subtypes 1a and 1b and the subtype 1a with NS3-K80 or NS3-Q80 polymorphism.
Owner:DIAGCOR LIFE SCI LTD
Efficient cell culture system for hepatitis c virus genotype 2b
InactiveUS20110294194A1High mutation rateReduce inhibitionSsRNA viruses positive-senseSugar derivativesSequence analysisHCV Genotyping
The present inventors developed hepatitis C virus 2b / 2a intergenotypic recombinants in which the JFH1 structural genes (Core, E1 and E2), p7 and the complete NS2 were replaced by the corresponding genes of the genotype 2b reference strain J8. Sequence analysis of recovered 2b / 2a recombinants from 2 transfection experiments revealed that 2b / 2a was genetically stable. Conclusion: The developed 2b / 2a viruses provide a robust in vitro tool for research in HCV genotype 2b, including vaccine studies and functional analyses.
Owner:HVIDOVRE HOSPITAL
Hepatitis-C virus testing
New styles of hepatitis C virus (HCV), referred to as HCV-3 and HCV-4, have been identified and sequenced. Antigenic regions of HCV-2, HCV-3 and HCV-4 polypeptides have been identified. Immunoassays for HCV and antibodies thereto are described, which allow more complete screening of blood samples for HCV, and allow HCV genotyping.
Owner:COMMON SERVICES AGENCY
HCV vaccines and methods for using the same
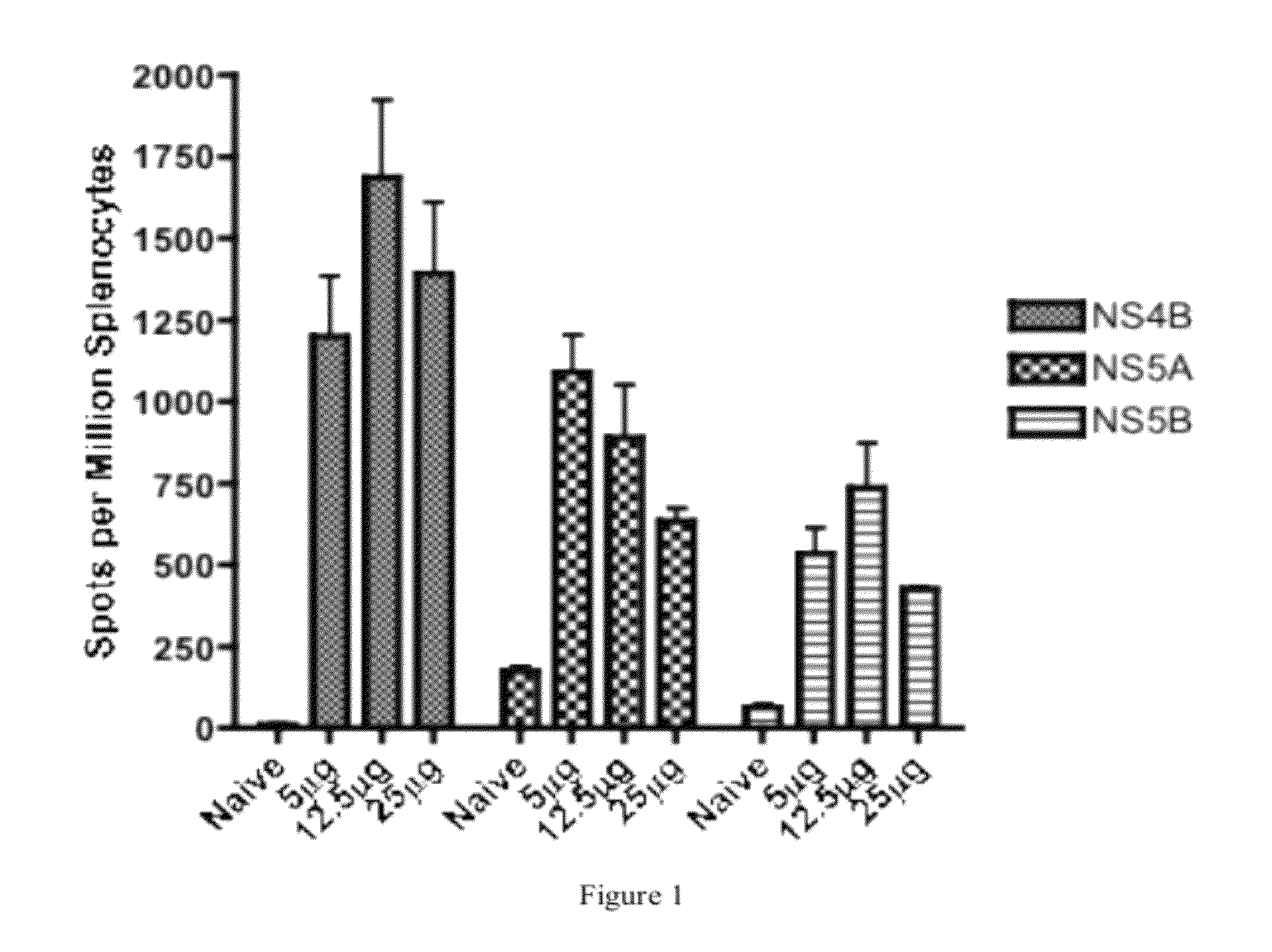
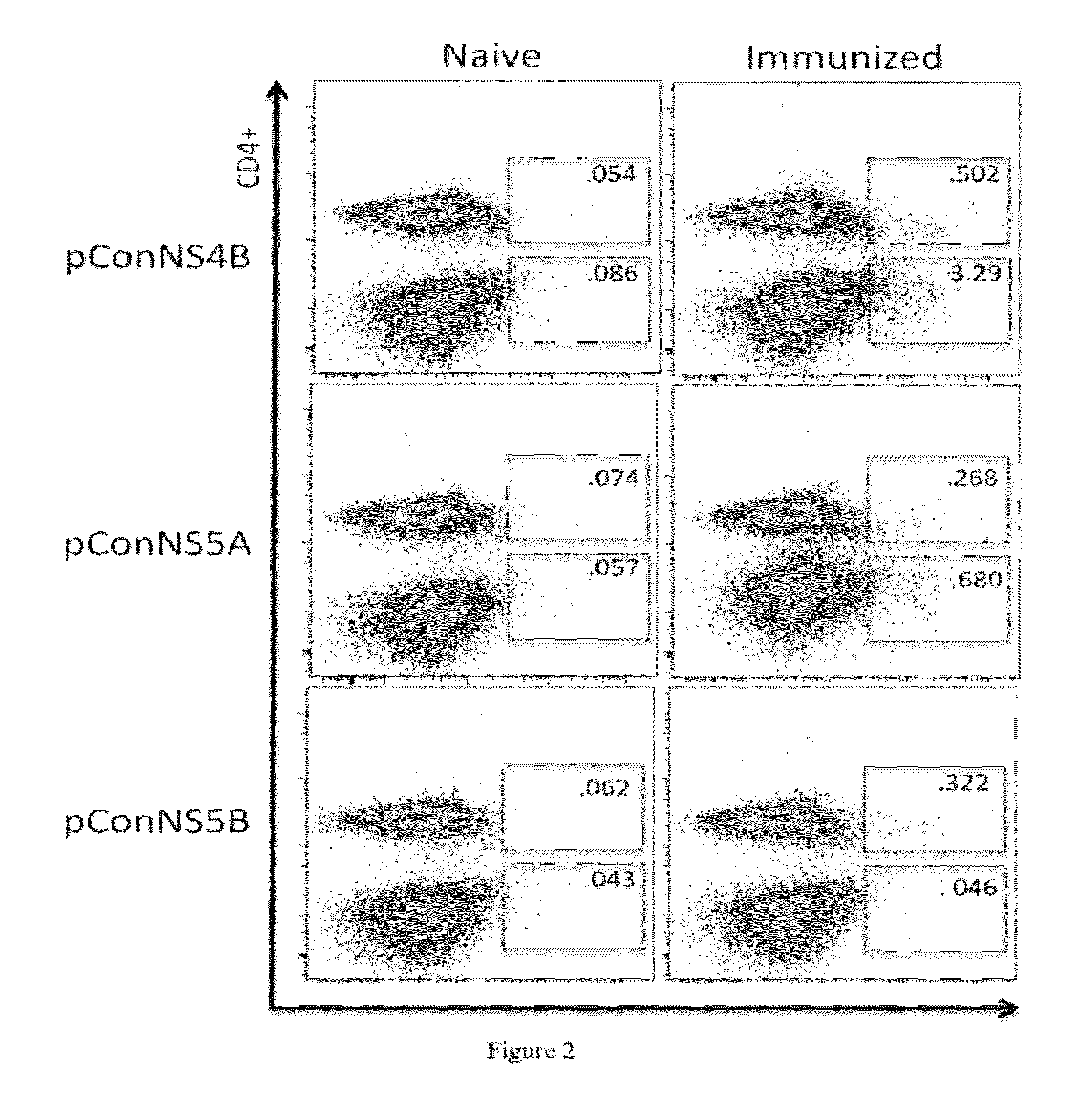
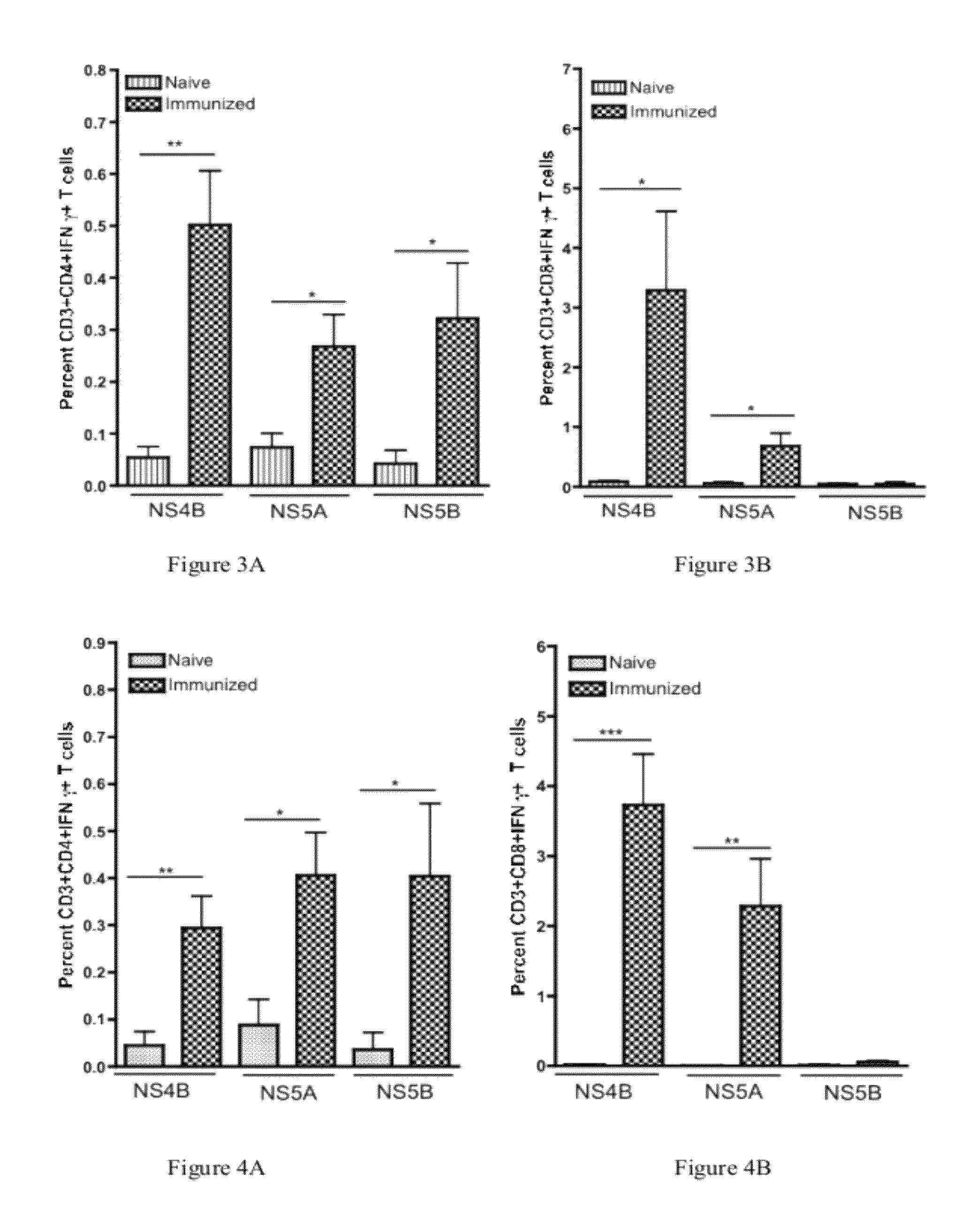
Improved anti-HCV immunogens and nucleic acid molecules that encode them are disclosed. Immunogens disclosed include those having consensus HCV genotype 1a, including for example, NS4B, NS5A and NS5B. Pharmaceutical composition, recombinant vaccines comprising and live attenuated vaccines are disclosed as well methods of inducing an immune response in an individual against HCV are disclosed.
Owner:THE TRUSTEES OF THE UNIV OF PENNSYLVANIA +1
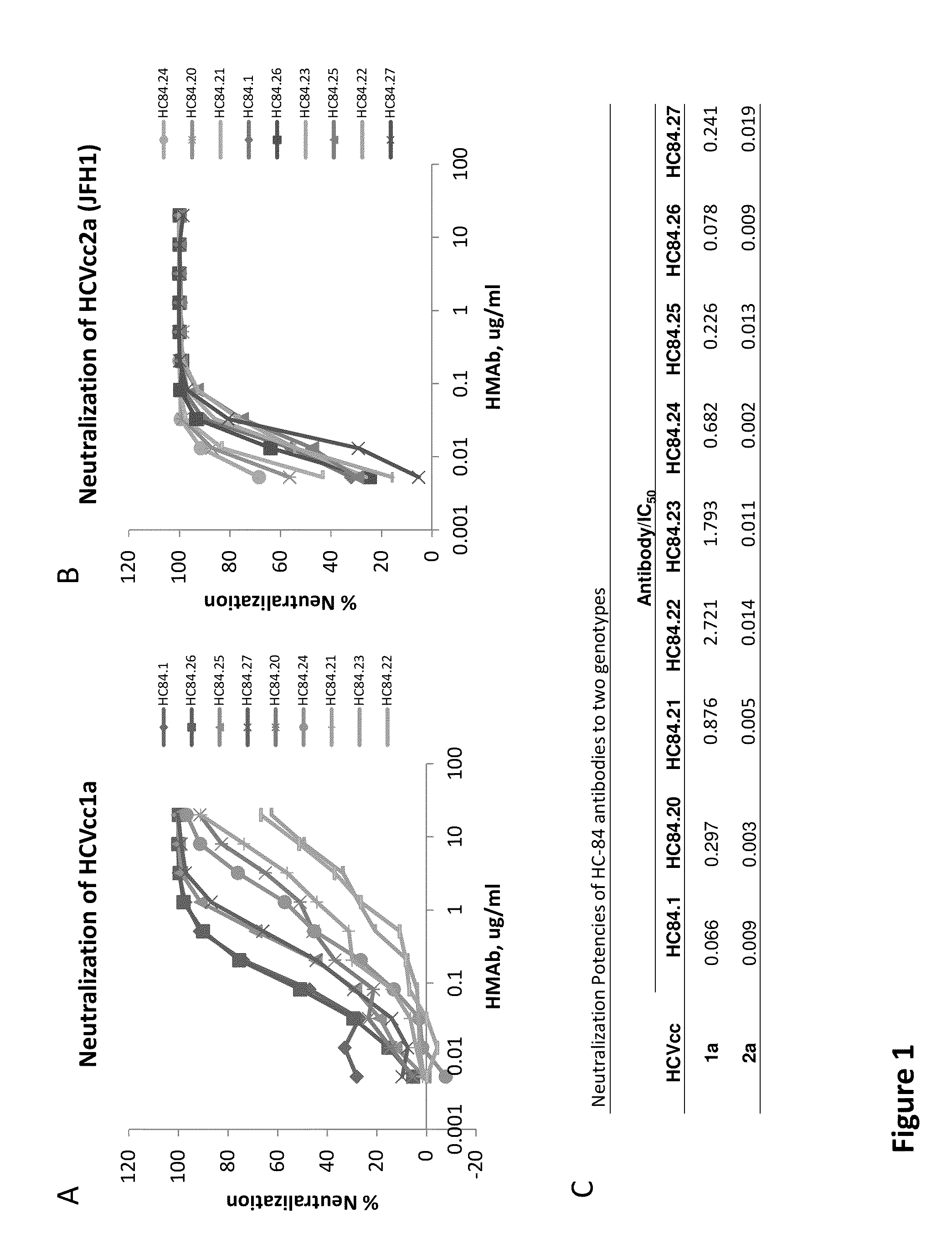
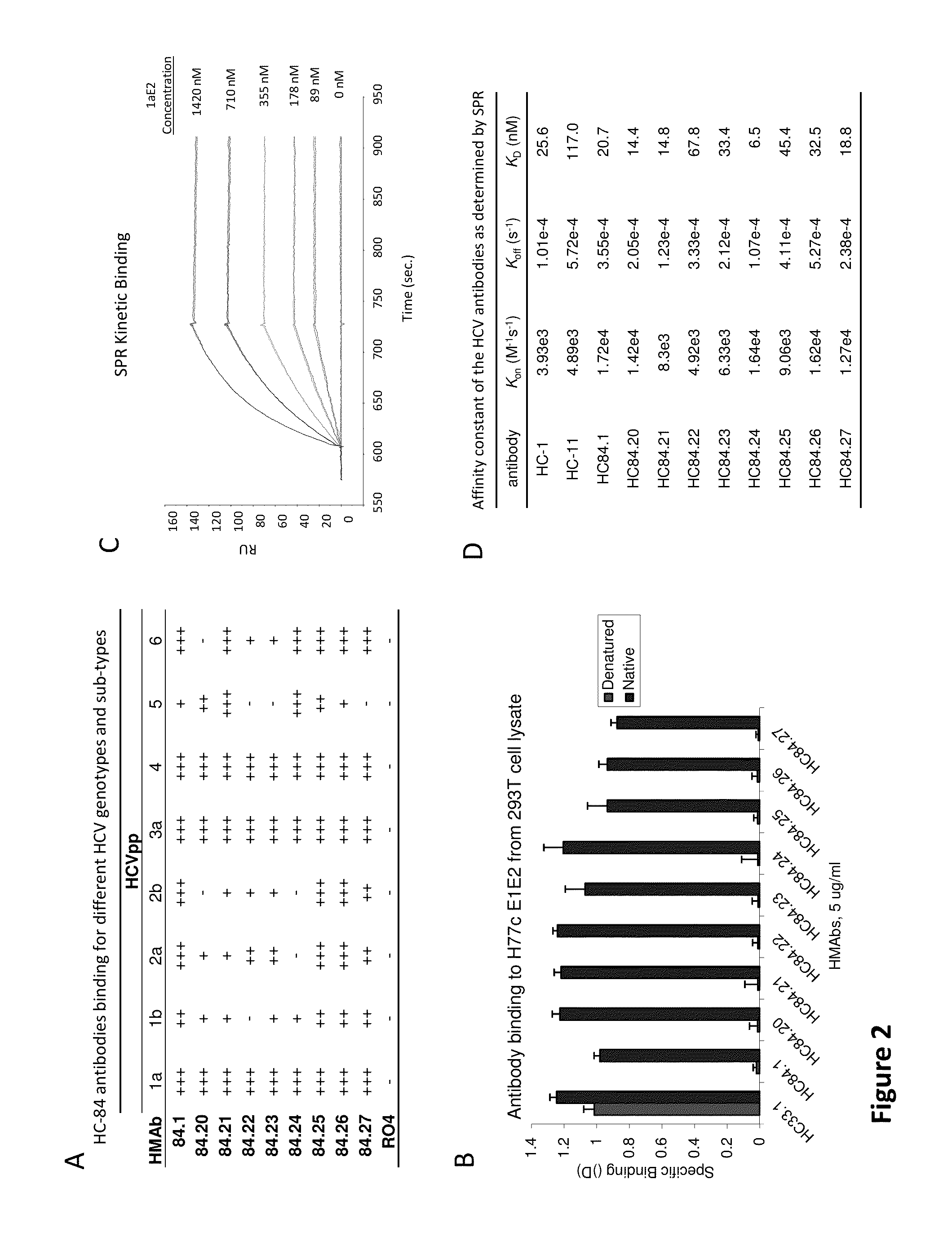
Cluster of Neutralizing Antibodies to Hepatitis C Virus
InactiveUS20130084301A1Reduce decreaseAnimal cellsSsRNA viruses positive-senseHCV GenotypingMonoclonal antibody
Compositions and methods are provided relating to human anti-HCV E2 monoclonal antibodies. The antibodies of the invention bind to a conserved region of HCV E2 protein, and neutralize HCV influenza virus across multiple HCV genotypes. Embodiments of the invention include isolated antibodies and derivatives and fragments thereof, pharmaceutical formulations comprising one or more of the human anti-HCV monoclonal antibodies; and cell lines that produce these monoclonal antibodies.
Owner:THE BOARD OF TRUSTEES OF THE LELAND STANFORD JUNIOR UNIV
Combination Therapy Regimen For Treatment Of Selected HCV Genotypes
InactiveUS20180021360A1Lower potentialMinimizes potential for drug toxicityPowder deliveryOrganic active ingredientsHCV GenotypingFixed dose
A method for the treatment of hepatitis C infection genotype 1, 2, 4, 5, or 6, but not genotype 3 is provided comprising administering an effective amount of a combination of Compound (I), Compound (II), and Compound (III), or independently optionally their pharmaceutically acceptable salt, solvate or hydrate, optionally in a solid fixed dose composition.
Owner:JANSSEN PHARMA INC +1
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com