Nucleotide sequence and kit for detecting hepatitis C viruses
A hepatitis C virus, nucleic acid sequence technology, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem that the performance indicators cannot reach clinical diagnosis and treatment, and the detection sensitivity and stability are poor. , unable to meet the detection needs and other problems, to achieve the effect of high specificity, high sensitivity and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
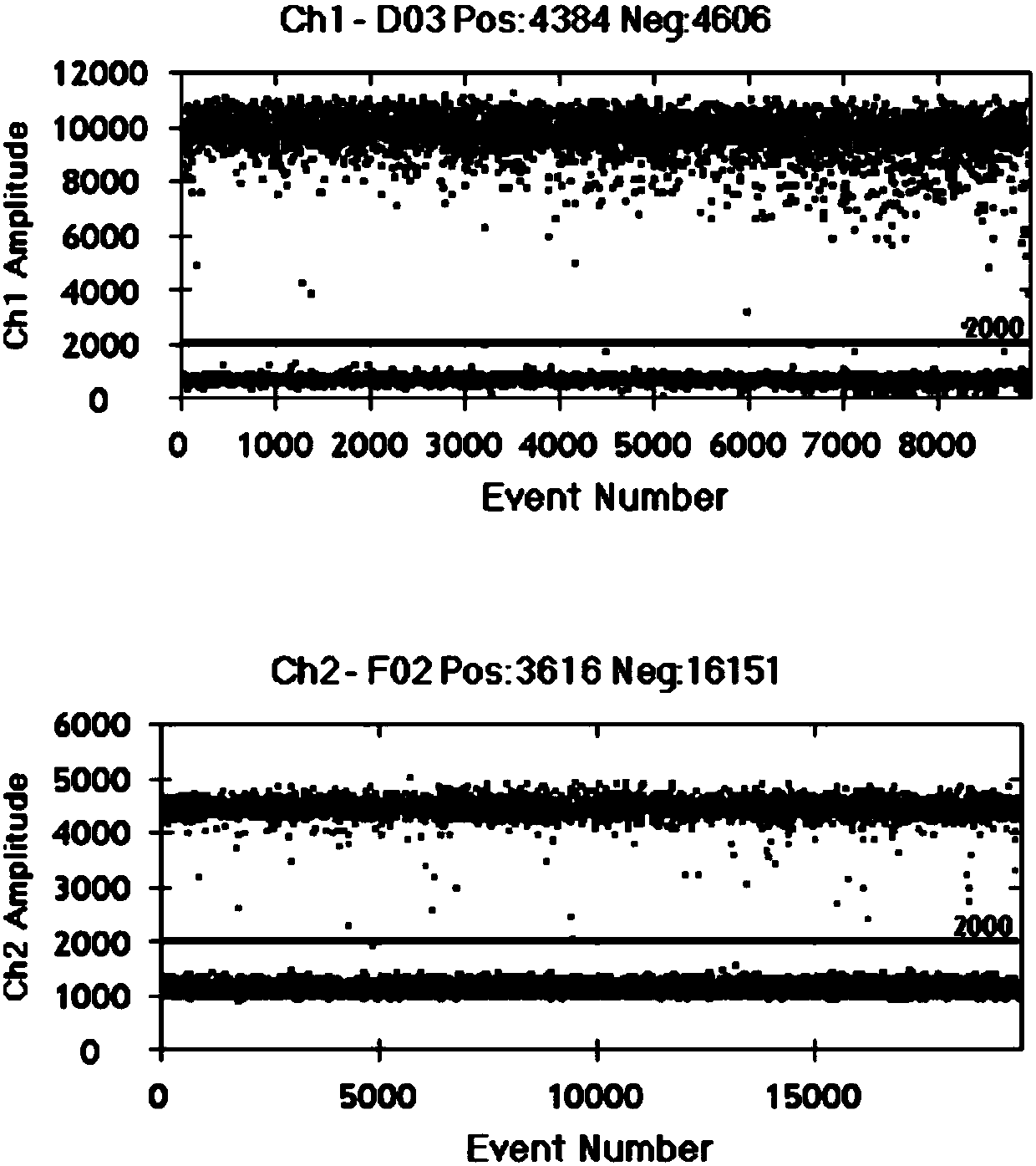
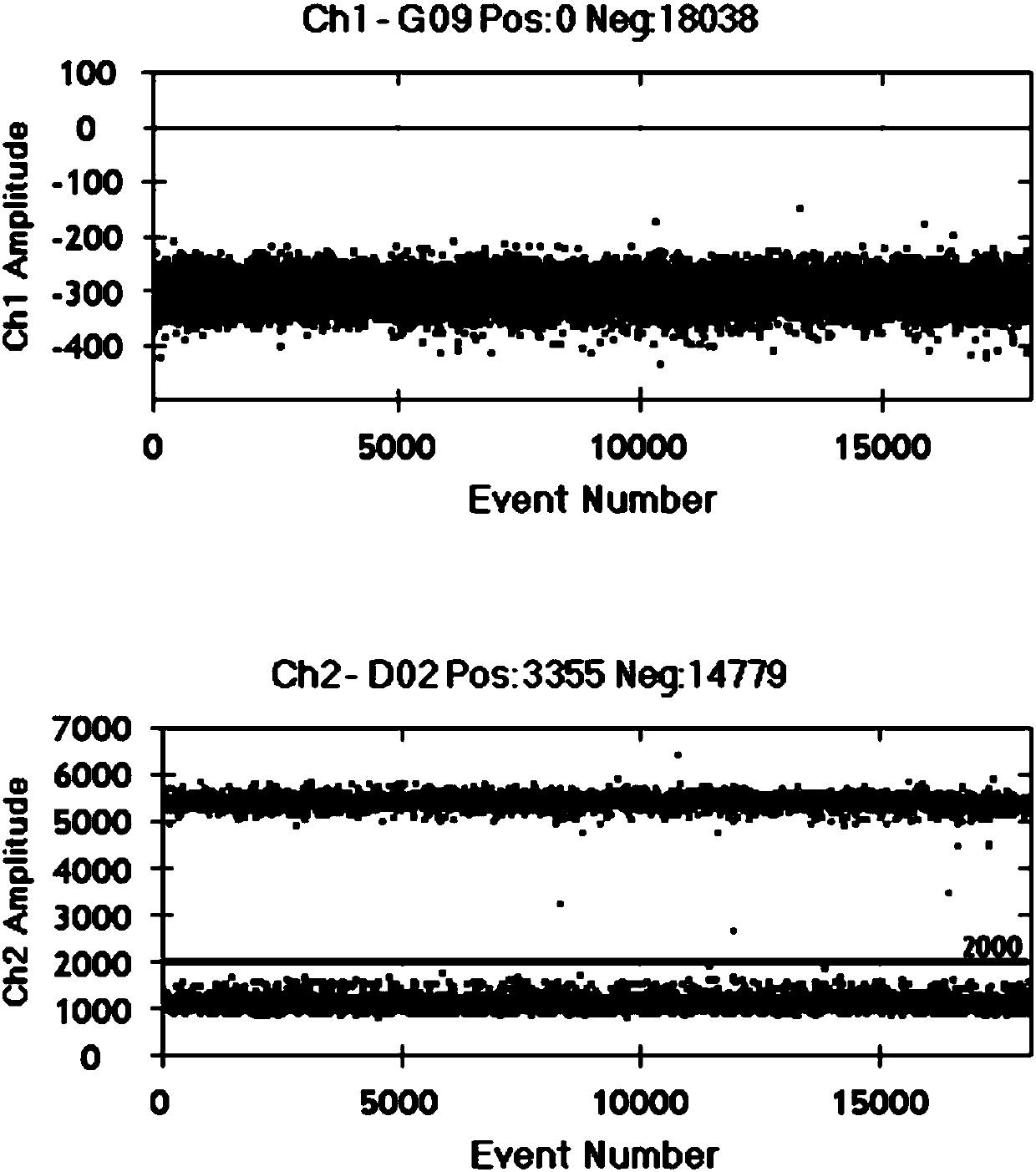
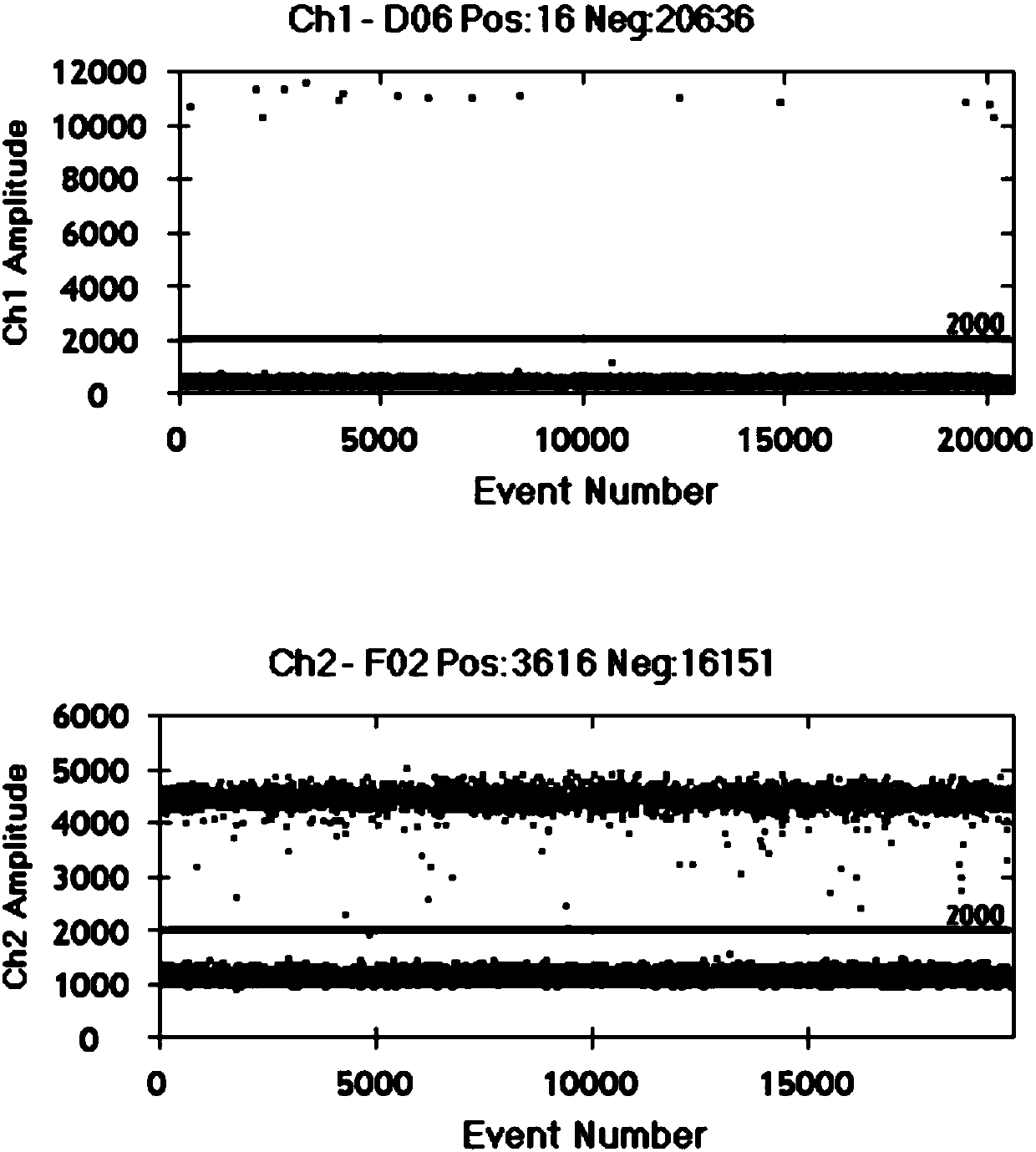
Image
Examples
Embodiment 1
[0036] Embodiment 1: The primers and probes that detect HCV RNA and human internal reference gene ERG are synthesized by Yingwei Jieji (Shanghai) Trading Co., Ltd., and the sequences are as follows:
[0037] Primers for amplifying HCV DNA:
[0038] F1 (SEQ ID NO: 1): 5'-ccgggagrgccatagtrgtct-3';
[0039] R1 (SEQ ID NO: 2): 5'-gtttatccaagaaaggacccdgtc-3';
[0040] Probes to detect HCV RNA:
[0041] PHCV (SEQ ID NO: 3): 5'-actcaccggttccg-3';
[0042] Wherein, r represents a degenerate base (a or g), and d represents a degenerate base (g or a or t); the 5' end of the HCV probe PHCV (SEQ ID NO: 3) sequence is marked with a FAM fluorescent group, The 3' end is labeled with the MGB quencher group,
[0043] Primers for amplifying the human ERG gene:
[0044] F3 (SEQ ID NO: 4): 5'-cagaacacagcagagggaagg-3';
[0045] R3 (SEQ ID NO: 5): 5'-aagctcccacttccataaaggc-3';
[0046] Probes for detection of human ERG gene:
[0047] PERG (SEQ ID NO: 6): 5'-aggagtcccaggctc-3';
[0048] Whe...
Embodiment 2
[0049] Example 2: Preparation method of HCV RNA quantitative kit.
[0050] (1) PCR reaction solution: 4*supermix (purchased from U.S. Bio-rad company, article number 1863026), is 4*PCR reaction premix, wherein contains PCR reaction buffer solution of the present invention, hot-start DNA polymerase, Mg 2+ and dNTPs and other components, stored at -20°C;
[0051] (2) Reverse Transcriltase: Store at -20°C
[0052] (3) DTT: 300nM: Store at -20°C
[0053] (4) Primer-probe premix solution: Submit the nucleotide sequences shown in SEQ ID NO: 1 to 6 to Yingwei Jieji (Shanghai) Trading Co., Ltd. for synthesis. After getting the dry powder, use deionized water to separate the Dilute 4 primers to a concentration of 100uM, dilute two probes to a concentration of 10uM, then mix the 6 primer-probe master solutions at a ratio of 9:9:25:9:9:25, and store at -20°C;
[0054] (5) Positive control: Serum samples containing HCV 7 genotype plasmids (purchased from the China Institute of Standard...
Embodiment 3
[0057] Embodiment 3: detection method.
[0058] Instruments: Eppendorf Mastercycler pro S qualitative PCR instrument, Bio-rad QX200 droplet digital PCR system (including droplet generator, sealing device, droplet reader), BECKMAN 22R desktop micro-refrigerated centrifuge, WH-866 vortex oscillator (Taicang Hualida), low-speed plate centrifuge (Anhui Zhongjia).
[0059] (1) Preparation of HCV patient samples, positive control, and negative control RNA templates: Use QIAGEN Serum Virus Extraction Kit (column method, Cat. No. 57704), operate according to the kit instructions, and extract patient samples into 500 μL serum in advance Add 2.5 μL of the internal reference plasmid, and then extract; take 500 μL of negative and positive control samples for extraction directly, and use 50 μL of eluent for elution, and use it as a PCR reaction template for later use.
[0060] (2) using the nucleic acid obtained in step (1) as a template, using 3 pairs of specific primers and two specifi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com