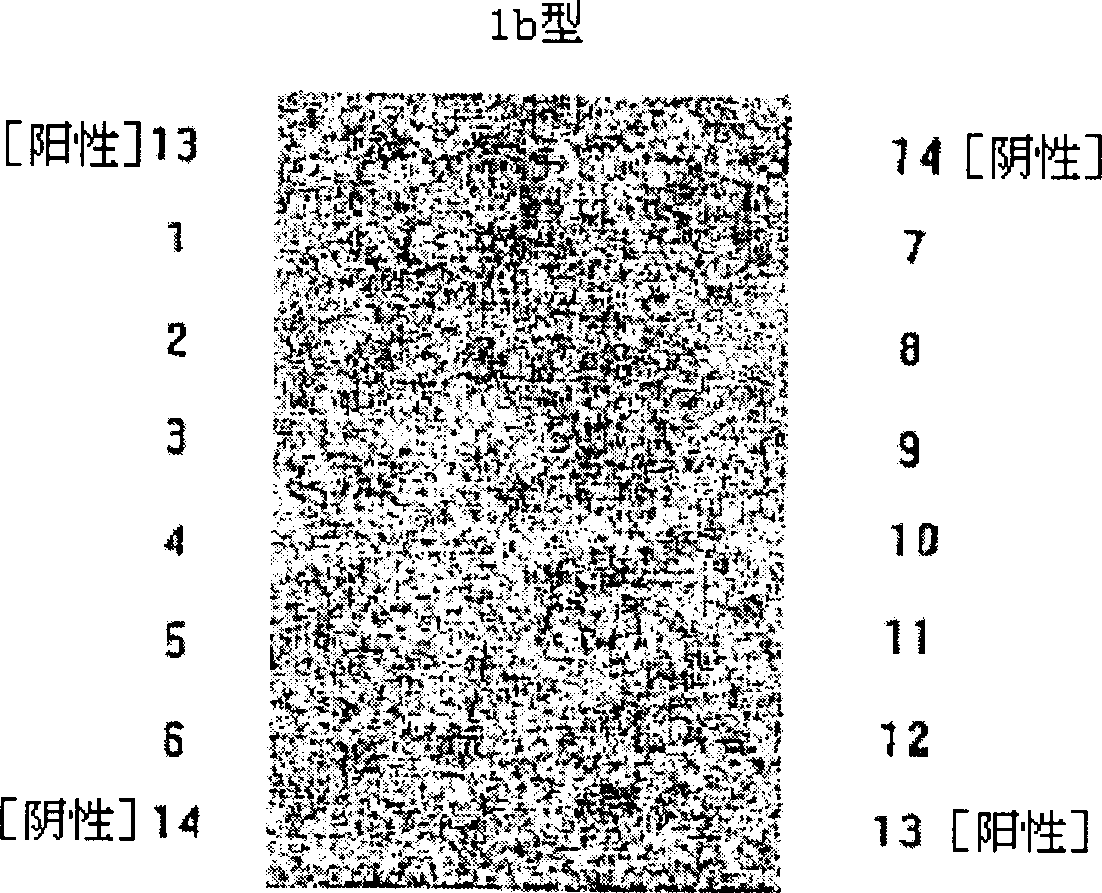
Oligonucleotides chip composition for analyzing HCV gene type and its detecting method
The technology of oligonucleotide probe and composition is applied in the field of oligonucleotide chip composition for analyzing hepatitis C virus genotype and its detection field, which can solve the problems of time-consuming and labor-intensive processing and analysis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Example 1: Artificial synthesis and base sequence of HCV primers
[0021] HCV PCR primers as shown in Table 1 were used for RT-PCR by analyzing the sites that co-react with the type 6 5'UTR. Antisense primers for secondary asymmetric PCR were classified into two categories according to the method of identification. In the first category, primers are attached to biotin to identify chromogenic reactions by hybridization. In the second class, anthocyanins were used to identify fluorescent reactions through hybridization reactions. An additional 25 bp base (SP6) was artificially synthesized at the 5' end of the secondary PCR antisense primer, and a base complementary to this site linked to anthocyanin was synthesized. The primers were synthesized by MWG-biotech company (Germany) according to the inventor's reservation, and the primers were synthesized by the method for synthesizing oligonucleotides written in Section 10.42 of the third edition of "Molecular Cloning" (Sa...
Embodiment 2
[0023] Embodiment 2: the preparation of HCV RNA, reverse transcription-primary PCR and secondary PCR reaction
[0024] 1) Mix 5 ul of HCV RNA extraction buffer (DEPC-DW 860 ul in 1 ml, Taq 10×buffer 100 ul, 1M DTT 20 ul, 10% NP40 20 ul) and 10 ul of plasma or serum.
[0025] 2) Heat the test tube containing the mixed serum in the PCR equipment at 92°C for 2 minutes, then quickly tap the test tube on ice
[0026] 3) Centrifuge the test tube for 5-10 seconds
[0027] 4) Then carry out reverse transcription and primary PCR reaction in GeneAmp PCR system 9600 thermal cycler (Perkin ElmerCetus, U.S.A) as shown in Table 2
[0028] 5) Carry out secondary asymmetric PCR reaction with 2ul primary PCR product as shown in Table 3
[0029] 6) Mix 1 ul of gel loading buffer (0.25% bromophenol blue, 0.25% xylidine FF, 15% non-polysucrose 400) into 5 ul of the secondary asymmetric PCR product, and add 1 μg / ml of bromine After electrophoresis on 2% agarose gel with ethidium (EtBr), a ban...
Embodiment 3
[0032] Embodiment 3: be used for the synthesis and base sequence of the probe of preparation HCV oligonucleotide chip
[0033] All probes have an amino linkage added to the 5' end for covalent attachment to acetaldehyde glass. 10-20 oligo(dT) are attached to the probe to facilitate the hybridization reaction. Then, the base sequence shown in Table 5 was ligated to amino-linked-oligo(dT) 10-20 . Short representation, with "amino linkage - oligo(dT) 10-20 -The primers of the "probe base sequence" arrangement order are synthesized in MWG-biotech company (Germany) according to the inventor's reservation. The base sequences of the identified HCV genotypes are analyzed as shown in Table 4 and belong to 6 types. HCV genes from 53 species were identified.
[0034] HCV type
HCV category
1a
HCV-1(M62321), HCV-H(M67463), HC-J1(D10749), GM1(M61728),
GM2(M61719), H90(M62382), US9(L38353), H77(AF009606), H99, PT-1
1b
HCV...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com