A kit for genotyping hepatitis C virus
A hepatitis C virus and genotyping technology, applied in the field of medical devices, can solve the problems of inability to meet clinical needs, low sensitivity, low specificity, insufficient subtyping ability, etc., achieving a high degree of automation, accurate results, Actionable fast effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Preparation of the kit of the invention
[0041] The composition of the kit of the present invention is as follows:
[0042] (1) Hepatitis C virus RNA extraction reagent: TRIzol lysate, chloroform, isopropanol and absolute ethanol
[0043] (2) Primer:
[0044] The sequence of the upstream primer of HCV NS5B gene is: 5’ -TTAACCACATCMRCTCCGTGTG-3’
[0045] The downstream primer sequence of HCV NS5B gene is: 5’-GTACCTGGTCATAGCYTCCGTRAA-3’
[0046] The sequence of the upstream primer of HCV NCR conserved region gene is: 5’-GCGGAACCGGTGAGTACA-3’
[0047] The downstream primer sequence of HCV NCR conserved region gene is: 5’-CCTATCAGGCAGTACCACAAGG-3’
[0048] The above primers were synthesized by Shanghai Life Technology Company.
[0049] (3) Negative control and positive control: deionized water is used as a negative control, and a sample containing HCV RNA is used as a positive control.
[0050] (4) Reverse transcription PCR reagents: 200 U / μL M-MLV, 40 U / μL RNase, 50 μM Oligo(...
Embodiment 2
[0057] Example 2 Using the kit prepared in Example 1 to detect the genotype of HCV
[0058] Take the detection of HCV genotype in peripheral blood samples of 20 hepatitis C patients as an example.
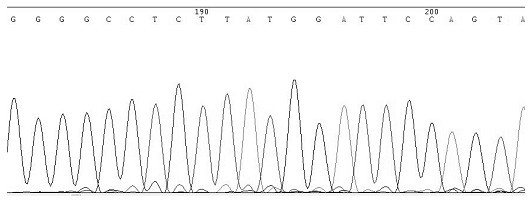
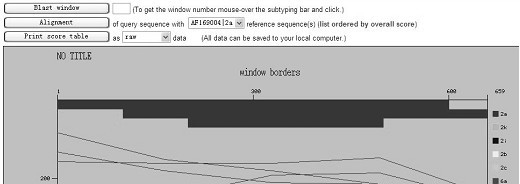
[0059] Detection process: First, design specific primers based on the HCV nucleic acid sequence provided by the HCV nucleic acid database. Obtain peripheral blood samples of clinical hepatitis C patients, quickly extract HCV RNA, use the extracted RNA as a template, and perform reverse transcription reaction with synthetic oligonucleotide reverse transcription primers to obtain cDNA, and then use this cDNA as a template and synthesis PCR primers are used for PCR amplification. The amplified product is directly recovered by the gel recovery kit for rapid gel recovery, the recovered DNA is used for the sequencing reaction, the sequencing reaction product is purified and then sequenced, and finally the nucleic acid sequence is compared in the NCBI nucleic acid database to determine the H...
Embodiment 3
[0070] Example 3 Evaluation of the detection capability of the kit of the present invention
[0071] According to Example 1, 45 specimens of HCV patients who were genotyped by restriction fragment length polymorphism method were tested using the kit of the present invention. The detection capabilities of the two were compared. The sensitivity and specificity of the kit were compared. Compared with the restriction fragment length polymorphism method for typing and sensitivity, this kit is more accurate and fully meets the current practical requirements of clinical diagnosis and treatment:
[0072]
[0073] among them:
[0074] ① Specificity: 100%;
[0075] ② Sensitivity: 97%;
[0076] ③ Positive predictive value: the positive predictive value reaches 100%;
[0077] ④ Negative predictive value: Negative predictive value reaches 94%;
[0078] ⑤ Repeatability: the results of repeated experiments are consistent;
[0079] ⑥ Time-consuming: The testing time for a clinical specimen is about 8-1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com